Epigenetic Inheritance in Rice Plants
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genotypic Performance, Adaptability and Stability in Special Types of Irrigated Rice Using Mixed Models1
Revista Ciência Agronômica, v. 50, n. 1, p. 66-75, jan-mar, 2019 Centro de Ciências Agrárias - Universidade Federal do Ceará, Fortaleza, CE Scientific Article www.ccarevista.ufc.br ISSN 1806-6690 Genotypic performance, adaptability and stability in special types of irrigated rice using mixed models1 Desempenho genotípico, adaptabilidade e estabilidade em tipos especiais de arroz irrigado via modelos mistos Eduardo Anibele Streck2*, Ariano Martins de Magalhães Júnior3, Gabriel Almeida Aguiar2, Paulo Henrique Karling Facchinello2 and Paulo Ricardo Reis Fagundes3 ABSTRACT - There is an increasing demand for special types of rice that have black or red pericarp, low amylose (subspecie japonica) or aromatic. However, intense breeding process for obtaining and indicating genetically promising new cultivars and adapted to cultivation environments are demanded. In this sense, for evaluating the special types of irrigated rice genotypes, and determining the adaptability and stability of these using mixed models. First, a preliminary agronomic evaluation of genotypes for special types of irrigated rice was performed, and later, the agronomically promising genotypes were evaluated in multi-site trials, aiming at verifying the interactions with the environment. Statistical analyzes were performed considering mixed models, using the SELEGEN-REML/BLUP software. High genotypic variability among genotypes for special types of irrigated rice was observed, obtaining some agronomically promising and with good adaptability and stability, with high accuracy and selection efficiency using mixed models. The observed wide diversity and rice genetic variability presented new prospects and opportunities for producers for acquiring food of higher added value to consumer market. Key words: Oryza sativa. Black rice. Red rice. Aromatic rice. -

Junmai Ginjo MOMOKAWA 90 Ruby PTS Junmai Ginjo Craft Can Mildly Sweet with Hints of Red Berries, Cherry, a Medium-Dry, Crisp Saké with Fresh Aromas of Guava and Mochi
CRAFT BREWED IN THE PACIFIC NW AMERICA’S ORIGINAL CRAFT BREWERY VEGAN-FRIENDLY | GLUTEN-FREE | KOSHER NEW MOMOKAWA PRODUCT - Junmai Ginjo MOMOKAWA 90 Ruby PTS Junmai Ginjo Craft Can Mildly sweet with hints of red berries, cherry, A medium-dry, crisp saké with fresh aromas of guava and mochi. Hints of tropical fruit, melon and grapes on the nose. 92 melon, green apple and anise and subtle hints PTS of citrus and honeydew. Polish 58% ABV 14.8% SMV -3 Polish 58% ABV 14.1% SMV +4 Rice Exclusive California Calrose Rice Exclusive California Calrose Yeast Proprietary Enjoy chilled Yeast Proprietary Enjoy chilled AWARD 90pts, Exceptional, Best Buy, Gold Medal AWARD 92pts, Great Value, Gold Medal - Ultimate Wine Challenge ‘19 - World Saké Challenge ‘17 UPC: 250ml 7 47846 12250 2 UPC: 750ML 7 47846 13720 9 MOMOKAWA MOMOKAWA Organic - Junmai Ginjo SILVER Silver - Junmai Ginjo Notes of refreshing melon and lime combine SILVER Light, crisp and dry mouthfeel with hints of SILVER with delicate pineapple and cola flavors. mineral and citrus. Green apple, melon and spice on the nose. Polish 58% ABV 14.5% SMV -2 Rice Exclusive California Calrose Polish 58% ABV 14.8% SMV +7 Yeast Proprietary Enjoy chilled Rice Exclusive California Calrose Yeast Proprietary Enjoy chilled USDA CERTIFIED ORGANIC AWARDS Silver Medal - London Saké Challenge ‘19; AWARD Silver Medal - Rodeo Uncorked Int’l Wine Competition ‘19 Silver Medal - Tokyo Saké Competition ‘19 UPC: 300ML 7 47846 40051 8, 750ML 7 47846 40050 1, UPC: 750ML 7 47846 12720 0 19.5 L Keg 7 47846 40052 5 MOMOKAWA MOMOKAWA 94 Diamond - Junmai Ginjo 94 PTS PTS Organic - Junmai Ginjo Nigori 92 Medium-dry and crisp with a balance of soft Rich and silky layers of coconut and cream with a PTS water notes and fall flavors of apple and pear. -

Discovering Sake
Discovering Sake Embassy of Japan https://www.facebook.com/EmbassyofJapaninIndia/ http://www.in.emb-japan.go.jp/itprtop_en/index.html What Makes Sake So Great Sake has played a central role in Japanese life and culture for the past 2,000 years, while the knowledge and techniques involved in sake brewing spread to every corner of the country. In fact, sake is such an integral part of the Japanese diet that, having some knowledge of it can add to one’s understanding of Japanese history, culture, and society. Made primarily from rice, sake is a fermented beverage. It is brewed using a micro-organism called koji, along with yeast. Sake’s alcohol content varies from 13% to 16%, in general. It takes pristine water to make sake, and brewers take advantage of the various kinds of natural water available in Japan to make only the best. There are many different varieties of sake, and it can be enjoyed either warm or chilled, depending on the season. Sake might be just about the best medicine for whatever ails you, as long as it’s consumed in moderation. So say, “Kampai!” Established in 1953, the Japan Sake and Shochu Makers Association, pursuant to the Japanese Act on Securing of Liquor Tax and on Liquor Business Associations, oversees the promotion, development and distribution of Sake and Shochu in Japan and around the world. We represent 1700 Sake brewers Japan Sake and Shochu Makers Association and Shochu distillers throughout Japan. Address: Nihon Shuzo Toranomon Building 2nd Fl 1-6-15 Nishi Shimbashi, Minato-ku, Tokyo http://www.japansake.or.jp/sake/english/ 1 Message from Miss SAKE Miss SAKE has worked as global ambassador to spread the greatness of SAKE and Japanese cultures since 2013, at home and abroad. -

QTL Analysis for Plant and Grain Characters of Sake-Brewing Rice Using a Doubled Haploid Population
Breeding Science 52 : 309-317 (2002) QTL Analysis for Plant and Grain Characters of Sake-brewing Rice Using a Doubled Haploid Population Shinya Yoshida*1), Masaru Ikegami1), Junko Kuze2), Keiko Sawada2), Zentaro Hashimoto3), Takashige Ishii2), Chiharu Nakamura3) and Osamu Kamijima2) 1) Hyogo Prefectural Research Center for Agriculture, Forestry and Fisheries, 1533 Minamino-oka, Befu, Kasai, Hyogo 679-0198, Japan 2) Laboratory of Plant Breeding, Faculty of Agriculture, Kobe University, 1-1 Rokkodai, Nada, Kobe, Hyogo 657-8501, Japan 3) Laboratory of Plant Genetics, Faculty of Agriculture, Kobe University, 1-1 Rokkodai, Nada, Kobe, Hyogo 657-8501, Japan Rice (Oryza sativa L.) varieties used for brewing sake Introduction are commonly characterized by traits such as large grain size with white-core (an opaque structure inside Rice (Oryza sativa L.) is one of the most important sta- the rice grain). A linkage map was constructed using ple food crops in the world. In Japan, rice is also used for the doubled haploid lines derived from the cross of Reiho (a production of Japanese rice wine, sake. Sake is brewed from cooking variety) and Yamada-nishiki (a sake-brewing steamed rice through fermentation by koji (Aspergillus variety). Random amplified polymorphic DNA (RAPD), oryzae) and yeast. Rice varieties used strictly for sake- amplified fragment length polymorphism (AFLP) and brewing are characterized by a larger grain size than that of simple sequence repeat (SSR) marker systems were em- ordinary cooking rice, and by the presence of an opaque ployed in QTL analysis. A total of 145 markers were structure named white-core located in the center of rice identified and mapped on rice chromosomes. -

HAKUTSURU Hakutsuru Nishiki* - Junmai Daiginjo Sho-Une* - Junmai Daiginjo a Luxurious Saké, Mouth-filling and Full of flavor, Yet Delicately Refined
EXCLUSIVE JAPANESE IMPORTS HAKUTSURU 96 PTS Driven to create a one 94 of a kind saké rice strain PTS to rival Yamada-Nishiki SILVER (“the king of rice”), Hakutsuru spent over 10 years developing their own exceptional HAKUTSURU NISHIKI rice, becoming the first successful crossbreed of Yamadaho and Wataribune in 70 years. HAKUTSURU HAKUTSURU Hakutsuru Nishiki* - Junmai Daiginjo Sho-Une* - Junmai Daiginjo A luxurious saké, mouth-filling and full of flavor, yet delicately refined. Sho-Une, meaning “soaring clouds,” uses only the finest rice and Enjoy the elegance of refreshing floral notes of jasmine and orange renowned water. Delicate notes of apple and pear balance with lush blossom and savor a hint of vanilla sweetness in the extra-long finish. strawberry and nectarine. Polish 50% ABV 15.5% SMV +4 Polish 50% ABV 15.5% SMV +2 Rice Hakutsuru Nishiki Rice Yamada Nishiki Yeast Proprietary Yeast Proprietary Enjoy chilled Enjoy chilled AWARD 96pts - Tasting Panel ‘19 AWARDS 94pts, Gold Medal - World Saké Challenge ‘19; UPC: *720ML (DI or for Select States) 7 47846 06720 9 Silver Medal - S.F. Int’l Wine Competition ‘18 UPC: 300ML 7 47846 01010 6, 720ML 7 47846 01011 3, *1.8L (DI Only) 7 47846 01012 0 HAKUTSURU HAKUTSURU Sayuri - Nigori CLASS Superior - Junmai Ginjo CHAMP 92 Sayuri means “little lily” in Japanese, apt for this soft, PTS EXCEP- SILVER floral noted nigori. Hints of white grape and elements TIONAL A flowery and fragrant saké with silky, well-balanced of cherry blossom create a lush, creamy saké. smoothness. Notes of blueberry and melon with well 89 BEST PTS. -

Winter Delights
WINTER DEC–MAR 2018/19 No. 220 Free WESTERN JAPAN’S PREMIER VISITOR’S GUIDE Winter Delights Sake, skiing, spas and snowcrab Plus Inside… WHAT’S ON Explore Sights Food & Drink Discover EAT & DRINK A PERFECT LONG BEST SPOTS HOW SAKE IS KINOSAKI’S SEVEN SIGHTSEEING WEEKEND IN NARA FOR WINTER MAKING ITS HEALING SPRINGS SNOWSCAPES COMEBACK MAPS Discover Hiroshima’s Scenic Beauty YOICHI SHIDAREZAKURA CHERRY BLOSSOM A stunning 12m tall weeping cherry sitting atop a small in Akiota-cho hillock is lit up during its brief, but spectacular spring bloom OSORAKAN SNOW PARK Japan’s southernmost ski resort, offering long runs for all Breathtaking views, fabulous levels of skiers and snowboarders, as well as snow trekking and igloo building hiking, exciting snow sports and relaxing hot spring accommodation await SANDANKYO GORGE A stunning 13km river gorge, awarded 3 stars by the French travel guide Blue Guide and selected as one of the top 100 scenic beauty spots in Japan INI TANADA Picture-perfect terraced paddy fi elds, offering stunning photo opportunities year-round Nukui Springs Hotel Offers traditional outdoor hot spring baths overlooking Lake Ryuki in Kyoto Hiroshima’s Akiota-cho. Relax in comfortable western style or Japanese Himeji Okayama Osaka tatami mat rooms and enjoy our award-winning French dining. Akiota-cho Kobe Hiroshima Address: 4692-7, Kake, Akiotacho, Yamgatagun, Hiroshima 731-3501, Japan Tel: +81 (0)826-22-1200 Access: 1 hour by car from Hiroshima IC RESERVATIONS www.nukui-sp.com CONTENTS 26 Winter Dec / Jan / Feb 10 30 Features KANSAI FINDER 08 23 32 Events & Festivals Kansai’s Snowy Vistas Nara’s Cocktail Kings Where to experience the best of the region’s RASTA MUSICK winter snowscapes. -

Heap: a Highly Sensitive and Accurate SNP Detection Tool for Low-Coverage High-Throughput Sequencing Data
Heap: a highly sensitive and accurate SNP detection tool for low-coverage high-throughput sequencing data Item Type Article Authors Kobayashi, Masaaki; Ohyanagi, Hajime; Takanashi, Hideki; Asano, Satomi; Kudo, Toru; Kajiya-Kanegae, Hiromi; Nagano, Atsushi J.; Tainaka, Hitoshi; Tokunaga, Tsuyoshi; Sazuka, Takashi; Iwata, Hiroyoshi; Tsutsumi, Nobuhiro; Yano, Kentaro Citation Kobayashi M, Ohyanagi H, Takanashi H, Asano S, Kudo T, et al. (2017) Heap: a highly sensitive and accurate SNP detection tool for low-coverage high-throughput sequencing data. DNA Research. Available: http://dx.doi.org/10.1093/dnares/dsx012. Eprint version Publisher's Version/PDF DOI 10.1093/dnares/dsx012 Publisher Oxford University Press (OUP) Journal DNA Research Rights This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0/), which permits non-commercial re-use, distribution, and reproduction in any medium, provided the original work is properly cited. For commercial re-use, please contact [email protected] Download date 24/09/2021 06:06:16 Item License http://creativecommons.org/licenses/by-nc/4.0/ Link to Item http://hdl.handle.net/10754/623635 Dna Research, 2017, 0(0), 1–9 doi: 10.1093/dnares/dsx012 Full paper Full paper Heap: a highly sensitive and accurate SNP detection tool for low-coverage high-throughput sequencing data Masaaki Kobayashi1, Hajime Ohyanagi1,2, Hideki Takanashi3, Satomi Asano1, Toru Kudo1, Hiromi Kajiya-Kanegae3, Atsushi -
Details of Items
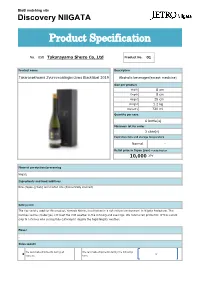
BtoB matching site Discovery NIIGATA Product Specification No. 058 Takarayama Shuzo Co,.Ltd Product No. 01 Product name Description Takaranokiwami Zyunnmaidaiginnzyou Blacklabel 2019 Alcoholic beverages(except medicine) Size per product Width 8 cm Depth 8 cm Height 29 cm Weight 1.2 kg Capacity 720 ml Quantity per case 6 bottle(s) Minimum lot for order 3 case(s) Expiration date and storage temperature Normal - Retail price in Japan (yen) *excluding tax 10,000 JPY Place of production/processing Niigata Ingredients and food additives Rice (Japan-grown) and malted rice (domestically sourced) Selling point The rice variety used for this product, Yamada Nishiki, is cultivated in a rich natural environment in Niigata Prefecture. This involves various challenges, not least the cold weather in the morning and evenings. We outsourced production of this superb crop to a farmer who successfully cultivated it despite the frigid Niigata weather. Flavor - Sales season We can make shipments during all We can make shipments during the following # ● # ~ seasons. term. BtoB matching site Discovery NIIGATA Product Specification No. 058 Takarayama Shuzo Co,.Ltd Product No. 02 Product name Description Takaranokiwami Zyunnmaidaiginnzyou Burgundylabel 2018 Alcoholic beverages(except medicine) Size per product Width 8 cm Depth 8 cm Height 29 cm Weight 1.2 kg Capacity 720 ml Quantity per case 6 bottle(s) Minimum lot for order 3 case(s) Expiration date and storage temperature Normal - Retail price in Japan (yen) *excluding tax 10,000 JPY Place of production/processing Niigata Ingredients and food additives Rice (Japan-grown) and malted rice (domestically sourced) Selling point Takara Shuzo leverages all the technology at his disposal for this masterpiece. -

BÂO Wine Menu Draft FINAL
SOUPS AND SALADS ENTREÉS LUST FOR LYCHEE V 13 VEGAN KHAO XOI V GF 17 savoy, radicchio, pistachio, goji dust, coconut butternut, coconut curry, rice noodles, carrot, bacon, everything wonton chips, chioggia beet shiitake, pickled mustard greens, chili dust chips and lychee vinaigrette SOFT SHELL CRAB FRIED RICE SOUTH CHINA SEA SALAD GF 19 fried soft shell crab, lump crab, red rice, 29 TEA glass noodles, savoy, cucumber, pickled shiitake, roasted corn, kombu, red onion, egg kombu, hijiki, smoked trout roe, crispy garlic ROOM Add Hamachi* GF +14 SCALLOP XO seared scallops, XO noodles, roasted root 45 at SOUP of the DAY MP vegetables, daikon, kimchi consomme YANG KWEI FEI* Bao five spice smoked duck breast, pork belly, 29 DIM SUM shiitake, onions, chilis, rice noodles, whiskey Brewhouse SMOKED YELLOWTAIL 17 oyster sauce onion, carrot, celery, served with radishes, roujiamo crustini SHORT RIB FRIED RICE GF braised short rib, shiitake, carrot, onion, 7 EVERYTHING CRAB RANGOON 17 2 garlic, ginger, egg, short rib jus jumbo lump crab, charred green onion infused cream cheese, fried everything SUGAR SESAME KOBE STEAK 6oz/10oz* GF mango wonton, spicy mustard sugar sesame crusted kobe beef steak, 55/85 roasted root vegetables, herb gremolata KOBE SHORT RIB DUMPLINGS 20 braised kobe beef, short rib and jus HAMACHI CRUDO* 17 hamachi, citrus, smoked chili salt and SHARED ENTREÉS 35/65 fresno oil PEKING DUCK Half/Whole 15 crispy duck, mandarin spring onion MILLION DOLLAR EGG ROLL* pancakes, plum sauce savoy, shiitake, spring onion, carrot, sprouts, radicchio, truffle, lime, cured egg WHOLE STEAMED FISH MP yolk fish of the day, black bean chili sauce, 13 spring onions, garlic, ginger LAO TAO five spice pork belly, spring onions, pork WHOLE FRIED FISH jus 45 fried Colorado raised rainbow trout, 21 crispy chilis, spring onions, radish, crispy LOBSTER CEVICHE garlic, chili oil fresh Lobster salad, celery, lettuce, rice wrapper, chilled 30 oz. -

Born in Japan, Made in Japan
MADE IN JAPAN, BORN IN JAPAN, EATEN GLOBALLY, E.A.K. Ramen aka Machida Shoten in Japan, started back in 2008 in Machida City, Japan. This was our rst shop to fulll our goal to spread the IEKEI style of ramen to the world. Our purpose was to be a shop that is loved by everyone, but especially locals. We wanted to provide the best possible ramen, service and atmosphere to our neighbors. With this in mind, we started our mission to spread IEKEI to the world. Iekei (pronounced E-A-K) is a style of ramen that originated from Yokohama Japan.It is a perfect balance of the popular Tonkotsu style of Kyushu in the West, and Shoyu style from Tokyo in the East. What makes our ramen distinct is that we use thick straight noodles to compliment the savory broth. Our EAK style is a modern take on the original iekei style. We’re delighted to bring this style of ramen to you. Don’t be afraid to go nuts on this bowl of ramen and slurp. Currently, we operate throughout Japan, Asia and parts of Europe. After capturing a major part of Japan, we decided the time was right to cross over the Pacic and enter the US market. To slurp? or 拉麺 not to slurp? RAMEN★= Popular It’s okay either way! ♥ = Recommended = Vegan SHOYU 元祖家系 SHIO 塩バター ★♥The E.A.K.醤 Ramen 15 Zebra Ramen 16 Our signature dish. Pork and chicken broth. Pork and chicken塩 broth, Shio tare and thick noodles. Shoyu tare and thick noodles. -
The Reverse Phenomenon in Water Absorption Rate in Japonica Rice Grains Caused by Long-Term Soaking in Water at Different Temperatures
日本調理科学会誌(J.Water Absorption Cookery Rates Sci. of Jpn.) Vol.Rice at Different 52,No. Soaking 3,159~168(2019) Temperatures〔Note〕 The Reverse Phenomenon in Water Absorption Rate in Japonica Rice Grains Caused by Long-term Soaking in Water at Different Temperatures Saeko Morii*,**,*** Kaoru Sakamoto*,** Naoko Shirasugi(Kataoka)*** § We investigated the influence of soaking temperature on the water absorption in three cultivars of Japonica rice, Kinuhikari, Sasanishiki, and Hatsushimo, based on the modified water absorption rate calculation method for rice. The grains of each cultivar were processed under different conditions, i.e., 70% polished and polished rice. We com- pared the water absorption curves of grains soaked in water at 5℃, 20℃, and 40℃ for 10 min to 24 h to ensure that the absorption had reached equilibrium. The absorption curves of grains soaked at 5℃ and 20℃ intersected those of grains soaked at 40℃ in all cultivars of both 70% polished rice and polished rice. This “reverse phenomenon” was specific to the granular shape of the rice grain. The findings for these three cultivars descended from the different rice lines contradict the established theories in the field of cookery science. It is suggested that this occurs for many cultivars of Japonica rice. Keyword:Japonica rice, polished rice, soaking temperature, water absorption and Adachi, 1992). The water absorption has also been INTRODUCTION reported to increase when the rice is soaked in water for Japonica rice, a staple of Japanese food, is shorter in a long time, even if the soaking temperature is low length and more viscous than Indica rice. -

Induced Mutations for Food and Energy Security: Challenge of Inducing Unique Mutants for New Cultivars and Molecular Research
農研機構報告 作物開発センター (Bull. NARO, Crop Sci.)1,33 - 124(2017) 33 Induced Mutations for Food and Energy Security: Challenge of Inducing Unique Mutants for New Cultivars and Molecular Research *1 Hitoshi NAKAGAWA and Hiroshi KATO Abstract Following the construction of the Gamma Field at the Institute of Radiation Breeding( IRB) in 1960, mutation breeding was accelerated in Japan. The facility is used to artifi cially induce mutations with a higher radiation dose( up to 2 Gy/day, that is ca. 300,000 times that of natural background) at a higher frequency than occurs in nature. Since the unit became operational, the number of mutant cultivars generated in Japan increased until 2000–2010 and has since decreased. There have been 295 direct-use mutant cultivars representing 70 species generated through irradiation utilizing gamma-rays, X-rays, ion beams and chemical mutagenesis and in vitro culture. Each cultivar has been registered and released in Japan, with approximately 79% of these induced by radiation. There have been 335 indirect-use mutant cultivars, including 298 rice, of which 150 cultivars(50 .3%) were derived from the semi-dwarf mutant cv. “Reimei” or its off spring. The economic impact of these mutant cultivars, primarily of rice and soybean, is very large. Some useful mutations are discussed for rice, such as low digestible-protein content, low amylose content, giant embryo and non-shattering. Useful mutations in soybean such as radio-sensitivity, fatty acid composition, lipoxygenase lacking, glycinin rich and super- nodulation have been identifi ed. A similar series of advantageous mutations have been found in Japanese pear and other crops through various screening methods.