Identification of Differentially Expressed Genes and Small Molecule Drugs for the Treatment of Tendinopathy Using Microarray Analysis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
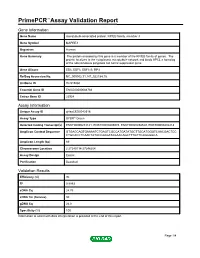
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name microtubule-associated protein, RP/EB family, member 3 Gene Symbol MAPRE3 Organism Human Gene Summary The protein encoded by this gene is a member of the RP/EB family of genes. The protein localizes to the cytoplasmic microtubule network and binds APCL a homolog of the adenomatous polyposis coli tumor suppressor gene. Gene Aliases EB3, EBF3, EBF3-S, RP3 RefSeq Accession No. NC_000002.11, NT_022184.15 UniGene ID Hs.515860 Ensembl Gene ID ENSG00000084764 Entrez Gene ID 22924 Assay Information Unique Assay ID qHsaCED0042518 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000233121, ENST00000405074, ENST00000458529, ENST00000402218 Amplicon Context Sequence GTGACCAGTGAAAATCTGAGTCGCCATGATATGCTTGCATGGGTCAACGACTCC CTGCACCTCAACTATACCAAGATAGAACAGCTTTGTTCAGGGGCA Amplicon Length (bp) 69 Chromosome Location 2:27245114-27246204 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 90 R2 0.9993 cDNA Cq 24.76 cDNA Tm (Celsius) 80 gDNA Cq 26.8 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report MAPRE3, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Human Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. -

EB3 (MAPRE3) (NM 012326) Human Tagged ORF Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RC204135 EB3 (MAPRE3) (NM_012326) Human Tagged ORF Clone Product data: Product Type: Expression Plasmids Product Name: EB3 (MAPRE3) (NM_012326) Human Tagged ORF Clone Tag: Myc-DDK Symbol: MAPRE3 Synonyms: EB3; EBF3; EBF3-S; RP3 Vector: pCMV6-Entry (PS100001) E. coli Selection: Kanamycin (25 ug/mL) Cell Selection: Neomycin ORF Nucleotide >RC204135 ORF sequence Sequence: Red=Cloning site Blue=ORF Green=Tags(s) TTTTGTAATACGACTCACTATAGGGCGGCCGGGAATTCGTCGACTGGATCCGGTACCGAGGAGATCTGCC GCCGCGATCGCC ATGGCCGTCAATGTGTACTCCACATCTGTGACCAGTGAAAATCTGAGTCGCCATGATATGCTTGCATGGG TCAACGACTCCCTGCACCTCAACTATACCAAGATAGAACAGCTTTGTTCAGGGGCAGCCTACTGCCAGTT CATGGACATGCTCTTCCCCGGCTGTGTGCACTTGAGGAAAGTGAAGTTCCAGGCCAAACTAGAGCATGAA TACATCCACAACTTCAAGGTGCTGCAAGCAGCTTTCAAGAAGATGGGTGTTGACAAAATCATTCCTGTAG AGAAATTAGTGAAAGGAAAATTCCAAGATAATTTTGAGTTTATTCAGTGGTTTAAGAAATTCTTTGACGC AAACTATGATGGAAAGGATTACAACCCTCTGCTGGCGCGGCAGGGCCAGGACGTAGCGCCACCTCCTAAC CCAGGTGATCAGATCTTCAACAAATCCAAGAAACTCATTGGCACAGCAGTTCCACAGAGGACGTCCCCCA CAGGCCCAAAAAACATGCAGACCTCTGGCCGGCTGAGCAATGTGGCCCCCCCCTGCATTCTCCGGAAGAA TCCTCCATCAGCCCGAAATGGCGGCCATGAGACTGATGCCCAAATTCTTGAACTCAACCAACAGCTGGTG GACTTGAAGCTGACAGTGGATGGGCTGGAGAAGGAACGTGACTTCTACTTCAGCAAACTTCGTGACATCG AGCTCATCTGCCAGGAGCATGAAAGTGAAAACAGCCCTGTTATCTCAGGCATCATTGGCATCCTCTATGC CACAGAGGAAGGATTCGCACCCCCTGAGGACGATGAGATTGAAGAGCATCAACAAGAAGACCAGGACGAG TAC ACGCGTACGCGGCCGCTCGAGCAGAAACTCATCTCAGAAGAGGATCTGGCAGCAAATGATATCCTGGATT -

Supporting Information
Supporting Information Pouryahya et al. SI Text Table S1 presents genes with the highest absolute value of Ricci curvature. We expect these genes to have significant contribution to the network’s robustness. Notably, the top two genes are TP53 (tumor protein 53) and YWHAG gene. TP53, also known as p53, it is a well known tumor suppressor gene known as the "guardian of the genome“ given the essential role it plays in genetic stability and prevention of cancer formation (1, 2). Mutations in this gene play a role in all stages of malignant transformation including tumor initiation, promotion, aggressiveness, and metastasis (3). Mutations of this gene are present in more than 50% of human cancers, making it the most common genetic event in human cancer (4, 5). Namely, p53 mutations play roles in leukemia, breast cancer, CNS cancers, and lung cancers, among many others (6–9). The YWHAG gene encodes the 14-3-3 protein gamma, a member of the 14-3-3 family proteins which are involved in many biological processes including signal transduction regulation, cell cycle pro- gression, apoptosis, cell adhesion and migration (10, 11). Notably, increased expression of 14-3-3 family proteins, including protein gamma, have been observed in a number of human cancers including lung and colorectal cancers, among others, suggesting a potential role as tumor oncogenes (12, 13). Furthermore, there is evidence that loss Fig. S1. The histogram of scalar Ricci curvature of 8240 genes. Most of the genes have negative scalar Ricci curvature (75%). TP53 and YWHAG have notably low of p53 function may result in upregulation of 14-3-3γ in lung cancer Ricci curvatures. -

In This Table Protein Name, Uniprot Code, Gene Name P-Value
Supplementary Table S1: In this table protein name, uniprot code, gene name p-value and Fold change (FC) for each comparison are shown, for 299 of the 301 significantly regulated proteins found in both comparisons (p-value<0.01, fold change (FC) >+/-0.37) ALS versus control and FTLD-U versus control. Two uncharacterized proteins have been excluded from this list Protein name Uniprot Gene name p value FC FTLD-U p value FC ALS FTLD-U ALS Cytochrome b-c1 complex P14927 UQCRB 1.534E-03 -1.591E+00 6.005E-04 -1.639E+00 subunit 7 NADH dehydrogenase O95182 NDUFA7 4.127E-04 -9.471E-01 3.467E-05 -1.643E+00 [ubiquinone] 1 alpha subcomplex subunit 7 NADH dehydrogenase O43678 NDUFA2 3.230E-04 -9.145E-01 2.113E-04 -1.450E+00 [ubiquinone] 1 alpha subcomplex subunit 2 NADH dehydrogenase O43920 NDUFS5 1.769E-04 -8.829E-01 3.235E-05 -1.007E+00 [ubiquinone] iron-sulfur protein 5 ARF GTPase-activating A0A0C4DGN6 GIT1 1.306E-03 -8.810E-01 1.115E-03 -7.228E-01 protein GIT1 Methylglutaconyl-CoA Q13825 AUH 6.097E-04 -7.666E-01 5.619E-06 -1.178E+00 hydratase, mitochondrial ADP/ATP translocase 1 P12235 SLC25A4 6.068E-03 -6.095E-01 3.595E-04 -1.011E+00 MIC J3QTA6 CHCHD6 1.090E-04 -5.913E-01 2.124E-03 -5.948E-01 MIC J3QTA6 CHCHD6 1.090E-04 -5.913E-01 2.124E-03 -5.948E-01 Protein kinase C and casein Q9BY11 PACSIN1 3.837E-03 -5.863E-01 3.680E-06 -1.824E+00 kinase substrate in neurons protein 1 Tubulin polymerization- O94811 TPPP 6.466E-03 -5.755E-01 6.943E-06 -1.169E+00 promoting protein MIC C9JRZ6 CHCHD3 2.912E-02 -6.187E-01 2.195E-03 -9.781E-01 Mitochondrial 2- -

Genetic Variation of SORBS1 Gene Is Associated with Glucose Homeostasis and Age at Onset of Diabetes: a Sapphire Cohort Study
www.nature.com/scientificreports OPEN Genetic variation of SORBS1 gene is associated with glucose homeostasis and age at onset of Received: 29 December 2017 Accepted: 19 June 2018 diabetes: A SAPPHIRe Cohort Study Published: xx xx xxxx Tien-Jyun Chang 1, Wen-Chang Wang2,3, Chao A. Hsiung3, Chih-Tsueng He4, Ming-Wei Lin5,6, Wayne Huey-Herng Sheu7,8,9, Yi-Cheng Chang1,10,11, Tom Quertermous12, Yii-Der Ida Chen13, Jerome I. Rotter13,14, Lee-Ming Chuang1,15 & The SAPPHIRe Study Group* The SORBS1 gene plays an important role in insulin signaling. We aimed to examine whether common single-nucleotide polymorphisms (SNPs) of SORBS1 are associated with prevalence and incidence of diabetes, age at onset of diabetes, and the related traits of glucose homeostasis. A total of 1135 siblings from 492 ethnic Chinese families were recruited at baseline, and 630 were followed up for 5.19 ± 0.96 years. Nine SNPs including rs7081076, rs2281939, rs3818540, rs2274490, rs61739184, rs726176, rs2296966, rs17849148, and rs3193970 were genotyped and examined. To deal with correlated data of subjects within the same families, the generalized estimating equations approach was applied throughout all association analyses. The GG genotype of rs2281939 was associated with a higher risk of diabetes at baseline, an earlier onset of diabetes, and higher steady-state plasma glucose levels in the modifed insulin suppression test. The minor allele T of rs2296966 was associated with higher prevalence and incidence of diabetes, an earlier onset of diabetes, and higher 2-h glucose during oral glucose tolerance test. These two SNPs revealed independent associations with age of diabetes onset as well as risk of diabetes at baseline. -

Vinexin Family (SORBS) Proteins Play Different Roles in Stiffness- Sensing and Contractile Force Generation Takafumi Ichikawa1,2, Masahiro Kita1, Tsubasa S
© 2017. Published by The Company of Biologists Ltd | Journal of Cell Science (2017) 130, 3517-3531 doi:10.1242/jcs.200691 RESEARCH ARTICLE Vinexin family (SORBS) proteins play different roles in stiffness- sensing and contractile force generation Takafumi Ichikawa1,2, Masahiro Kita1, Tsubasa S. Matsui3,4, Ayaka Ichikawa Nagasato1, Tomohiko Araki3, Shian-Huey Chiang5, Takuhito Sezaki1, Yasuhisa Kimura1, Kazumitsu Ueda1,2, Shinji Deguchi3,4, Alan R. Saltiel5,* and Noriyuki Kioka1,2,‡ ABSTRACT generating actin stress fibers (SFs) (Geiger et al., 2001). This Vinexin, c-Cbl associated protein (CAP) and Arg-binding protein 2 ‘ ’ (ArgBP2) constitute an adaptor protein family called the vinexin mechanical linkage acts as a molecular clutch to transmit the force (SORBS) family that is targeted to focal adhesions (FAs). Although derived from non-muscle myosin-II-dependent contraction to the numerous studies have focused on each of the SORBS proteins and ECM. Cells on more rigid substrates exert greater contractile forces partially elucidated their involvement in mechanotransduction, a than those on soft substrates (Hoffman et al., 2011; Roca-Cusachs comparative analysis of their function has not been well addressed. et al., 2012; LaCroix et al., 2015). These alterations can lead to Here, we established mouse embryonic fibroblasts that individually stiffness-dependent biochemical signals. Among the numerous FA scaffolding proteins, vinculin is one of expressed SORBS proteins and analysed their functions in an ‘ ’ identical cell context. Both vinexin-α and CAP co-localized with the main clutch molecules that can regulate force transmission. vinculin at FAs and promoted the appearance of vinculin-rich FAs, Vinculin consists of an N-terminal head region and a C-terminal tail α region separated by a flexible proline-rich linker region (Bakolitsa whereas ArgBP2 co-localized with -actinin at the proximal end of – FAs and punctate structures on actin stress fibers (SFs), and induced et al., 2004; Borgon et al., 2004). -

A Single Nucleotide Polymorphism in Exon 7 of Sorbin and SH3-Domain-Containing-1 (SORBS1) in Korean PCOS Patients
93-97 11/12/07 16:18 Page 93 MOLECULAR MEDICINE REPORTS 1: 93-97, 2008 93 A single nucleotide polymorphism in exon 7 of sorbin and SH3-domain-containing-1 (SORBS1) in Korean PCOS patients JUNG-MI PARK1, BON-HEE GU1, EUNG-JI LEE1, JAE-YOUNG KIM1, SUNG-WOO CHOI2 and KWANG-HYUN BAEK1 1Graduate School of Life Science and Biotechnology, Cell and Gene Therapy Research Institute, Pochon CHA University, CHA General Hospital, Seoul 135-081, Korea; 2Phillips Exeter Academy, Exeter, NH, USA Received October 19, 2007; Accepted November 20, 2007 Abstract. Patients with polycystic ovarian syndrome (PCOS) (3). It has been reported that approximately 4.9% of female are characterized by high levels of androgens, irregular or college students in Korea have exhibited PCOS (5). In women no menstrual cycle and increased hair growth. In addition, of other ethnic groups, that number has been reported as insulin resistance and glucose tolerance are caused in patients 4-12% (1,6,7). with PCOS. It was recently reported that PCOS in women is Recently, various single nucleotide polymorphisms (SNPs) associated with a single nucleotide polymorphism (SNP) of have been investigated by means of linkage and association genes, including the sorbin and SH3-domain-containing-1 studies in women with PCOS. A number of SNPs associated (SORBS1) gene involved in insulin resistance and glucose with PCOS have been reported (8-20), as have cases of PCOS uptake. SORBS1, which belongs to the sorbin homology related to the target genes of glucose uptake and insulin resis- (SoHo) family of proteins, becomes protein-protein interaction tance. -

Transcriptome Analysis of Adipose Tissues from Two Fat-Tailed Sheep
Li et al. BMC Genomics (2018) 19:338 https://doi.org/10.1186/s12864-018-4747-1 RESEARCHARTICLE Open Access Transcriptome analysis of adipose tissues from two fat-tailed sheep breeds reveals key genes involved in fat deposition Baojun Li1, Liying Qiao1, Lixia An2, Weiwei Wang1, Jianhua Liu1, Youshe Ren1, Yangyang Pan1, Jiongjie Jing1 and Wenzhong Liu1* Abstract Background: The level of fat deposition in carcass is a crucial factor influencing meat quality. Guangling Large-Tailed (GLT) and Small-Tailed Han (STH) sheep are important local Chinese fat-tailed breeds that show distinct patterns of fat depots. To gain a better understanding of fat deposition, transcriptome profiles were determined by RNA-sequencing of perirenal, subcutaneous, and tail fat tissues from both the sheep breeds. The common highly expressed genes (co-genes) in all the six tissues, and the genes that were differentially expressed (DE genes) between these two breeds in the corresponding tissues were analyzed. Results: Approximately 47 million clean reads were obtained for each sample, and a total of 17,267 genes were annotated. Of the 47 highly expressed co-genes, FABP4, ADIPOQ, FABP5,andCD36 were the four most highly transcribed genes among all the known genes related to adipose deposition. FHC, FHC-pseudogene,and ZC3H10 were also highly expressed genes and could, thus, have roles in fat deposition. A total of 2091, 4233, and 4131 DE genes were identified in the perirenal, subcutaneous, and tail fat tissues between the GLT and STH breeds, respectively. Gene Ontology (GO) analysis showed that some DE genes were associated with adipose metabolism. -

Views for Entrez
BASIC RESEARCH www.jasn.org Phosphoproteomic Analysis Reveals Regulatory Mechanisms at the Kidney Filtration Barrier †‡ †| Markus M. Rinschen,* Xiongwu Wu,§ Tim König, Trairak Pisitkun,¶ Henning Hagmann,* † † † Caroline Pahmeyer,* Tobias Lamkemeyer, Priyanka Kohli,* Nicole Schnell, †‡ †† ‡‡ Bernhard Schermer,* Stuart Dryer,** Bernard R. Brooks,§ Pedro Beltrao, †‡ Marcus Krueger,§§ Paul T. Brinkkoetter,* and Thomas Benzing* *Department of Internal Medicine II, Center for Molecular Medicine, †Cologne Excellence Cluster on Cellular Stress | Responses in Aging-Associated Diseases, ‡Systems Biology of Ageing Cologne, Institute for Genetics, University of Cologne, Cologne, Germany; §Laboratory of Computational Biology, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, Maryland; ¶Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand; **Department of Biology and Biochemistry, University of Houston, Houston, Texas; ††Division of Nephrology, Baylor College of Medicine, Houston, Texas; ‡‡European Molecular Biology Laboratory–European Bioinformatics Institute, Hinxton, Cambridge, United Kingdom; and §§Max Planck Institute for Heart and Lung Research, Bad Nauheim, Germany ABSTRACT Diseases of the kidney filtration barrier are a leading cause of ESRD. Most disorders affect the podocytes, polarized cells with a limited capacity for self-renewal that require tightly controlled signaling to maintain their integrity, viability, and function. Here, we provide an atlas of in vivo phosphorylated, glomerulus- expressed -

PRODUCT SPECIFICATION Product Datasheet
Product Datasheet QPrEST PRODUCT SPECIFICATION Product Name QPrEST MARE3 Mass Spectrometry Protein Standard Product Number QPrEST38970 Protein Name Microtubule-associated protein RP/EB family member 3 Uniprot ID Q9UPY8 Gene MAPRE3 Product Description Stable isotope-labeled standard for absolute protein quantification of Microtubule- associated protein RP/EB family member 3. Lys (13C and 15N) and Arg (13C and 15N) metabolically labeled recombinant human protein fragment. Application Absolute protein quantification using mass spectrometry Sequence (excluding YDGKDYNPLLARQGQDVAPPPNPGDQIFNKSKKLIGTAVPQRTSPTGPKN fusion tag) MQTSGRLSNVAPPCILRKNPPSARNGGHETDAQILELNQQLVDLKLTVDG LEKERDFYFSKLR Theoretical MW 30368 Da including N-terminal His6ABP fusion tag Fusion Tag A purification and quantification tag (QTag) consisting of a hexahistidine sequence followed by an Albumin Binding Protein (ABP) domain derived from Streptococcal Protein G. Expression Host Escherichia coli LysA ArgA BL21(DE3) Purification IMAC purification Purity >90% as determined by Bioanalyzer Protein 230 Purity Assay Isotopic Incorporation >99% Concentration >5 μM after reconstitution in 100 μl H20 Concentration Concentration determined by LC-MS/MS using a highly pure amino acid analyzed internal Determination reference (QTag), CV ≤10%. Amount >0.5 nmol per vial, two vials supplied. Formulation Lyophilized in 100 mM Tris-HCl 5% Trehalose, pH 8.0 Instructions for Spin vial before opening. Add 100 μL ultrapure H2O to the vial. Vortex thoroughly and spin Reconstitution down. For further dilution, see Application Protocol. Shipping Shipped at ambient temperature Storage Lyophilized product shall be stored at -20°C. See COA for expiry date. Reconstituted product can be stored at -20°C for up to 4 weeks. Avoid repeated freeze-thaw cycles. Notes For research use only Product of Sweden. -

Predicted Coronavirus Nsp5 Protease Cleavage Sites in The
bioRxiv preprint doi: https://doi.org/10.1101/2021.06.08.447224; this version posted June 8, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Predicted Coronavirus Nsp5 Protease Cleavage Sites in the 2 Human Proteome: A Resource for SARS-CoV-2 Research 3 Benjamin M. Scott1,2*, Vincent Lacasse3, Ditte G. Blom4, Peter D. Tonner5, Nikolaj S. Blom6 4 1 Associate, Biosystems and Biomaterials Division, National Institute of Standards and Technology, 5 Gaithersburg, Maryland, USA. 6 2 Department of Chemistry and Biochemistry, University of Maryland, College Park, Maryland, USA. 7 3 Segal Cancer Proteomics Centre, Lady Davis Institute, Jewish General Hospital, McGill University, Montreal, 8 Quebec, Canada 9 4 Department of Applied Mathematics and Computer Science, Technical University of Denmark, Lyngby, 10 Denmark. 11 5 Statistical Engineering Division, National Institute of Standards and Technology, Gaithersburg, Maryland, USA 12 6 Department of Bioengineering, Kongens Lyngby, Technical University of Denmark 13 14 *Corresponding author, [email protected] 15 16 BMS current affiliation: Concordia University, Centre for Applied Synthetic Biology, Montreal, Quebec, Canada 17 18 19 Abstract 20 Background: The coronavirus nonstructural protein 5 (Nsp5) is a cysteine protease required for 21 processing the viral polyprotein and is therefore crucial for viral replication. Nsp5 from several 22 coronaviruses have also been found to cleave host proteins, disrupting molecular pathways 23 involved in innate immunity. Nsp5 from the recently emerged SARS-CoV-2 virus interacts with 24 and can cleave human proteins, which may be relevant to the pathogenesis of COVID-19. -

Identification of Endogenous Adenomatous Polyposis Coli Interaction Partners and Β-Catenin−Independent Targets by Proteomics
Published OnlineFirst June 3, 2019; DOI: 10.1158/1541-7786.MCR-18-1154 Cancer Genes and Networks Molecular Cancer Research Identification of Endogenous Adenomatous Polyposis Coli Interaction Partners and b-Catenin– Independent Targets by Proteomics Olesja Popow1,2,3,Joao~ A. Paulo2, Michael H. Tatham4, Melanie S. Volk3, Alejandro Rojas-Fernandez5, Nicolas Loyer3, Ian P. Newton3, Jens Januschke3, Kevin M. Haigis1,6, and Inke Nathke€ 3 Abstract Adenomatous Polyposis Coli (APC) is the most frequently tion between endogenous MINK1 and APC and further mutated gene in colorectal cancer. APC negatively regulates confirmed the negative, and b-catenin–independent, regu- the Wnt signaling pathway by promoting the degradation of lation of MINK1 by APC. Increased Mink1/Msn levels were b-catenin, but the extent to which APC exerts Wnt/b-cate- also observed in mouse intestinal tissue and Drosophila nin–independent tumor-suppressive activity is unclear. To follicular cells expressing mutant Apc/APC when compared identify interaction partners and b-catenin–independent with wild-type tissue/cells. Collectively, our results highlight targets of endogenous, full-length APC, we applied label- the extent and importance of Wnt-independent APC func- free and multiplexed tandem mass tag-based mass spec- tions in epithelial biology and disease. trometry. Affinity enrichment-mass spectrometry identified more than 150 previously unidentified APC interaction Implications: The tumor-suppressive function of APC, the partners. Moreover, our global proteomic analysis revealed most frequently mutated gene in colorectal cancer, is mainly that roughly half of the protein expression changes that attributed to its role in b-catenin/Wnt signaling. Our study occur in response to APC loss are independent of b-catenin.