Identification of Endogenous Adenomatous Polyposis Coli Interaction Partners and Β-Catenin−Independent Targets by Proteomics
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Analyse Genetischer Stabilität in Den Nachkommen Bestrahlter Zellen Mittels Klassischer Chromosomenbänderung Und Verschiedener Hochdurchsatz-Techniken
Analyse genetischer Stabilität in den Nachkommen bestrahlter Zellen mittels klassischer Chromosomenbänderung und verschiedener Hochdurchsatz-Techniken Dissertation zur Erlangung des naturwissenschaftlichen Doktorgrades der Julius-Maximilians-Universität Würzburg vorgelegt von Julia Flunkert geboren in Schwerte Würzburg, 2018 Eingereicht am: …………………………………………….................... Mitglieder der Promotionskommission: Vorsitzender: Gutachter: Univ.-Prof. Dr. med. Thomas Haaf Gutachter: Univ.-Prof. Dr. Thomas Dandekar Tag des Promotionskolloquiums: ……………………….................. Doktorurkunde ausgehändigt am: ………………………................. Eidesstattliche Versicherung Die vorliegende Arbeit wurde von November 2014 bis September 2018 am Institut für Humangenetik der Universität Würzburg unter Betreuung von Herrn Univ.-Prof. Dr. med. Thomas Haaf angefertigt. Hiermit erkläre ich an Eides statt, die Dissertation: „Analyse genetischer Stabilität in den Nachkommen bestrahlter Zellen mittels klassischer Chromosomenbänderung und verschiedener Hochdurchsatz-Techniken“, eigenständig, d. h. insbesondere selbständig und ohne Hilfe eines kommerziellen Promotionsberaters, angefertigt und keine anderen, als die von mir angegebenen Quellen und Hilfsmittel verwendet zu haben. Ich erkläre außerdem, dass die Dissertation weder in gleicher noch in ähnlicher Form bereits in einem anderen Prüfungsverfahren vorgelegen hat. Weiterhin erkläre ich, dass bei allen Abbildungen und Texten bei denen die Verwer- tungsrechte (Copyright) nicht bei mir liegen, diese von den Rechtsinhabern -
Primepcr™Assay Validation Report
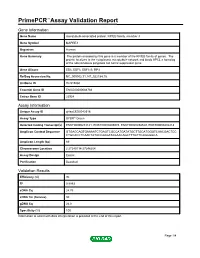
PrimePCR™Assay Validation Report Gene Information Gene Name microtubule-associated protein, RP/EB family, member 3 Gene Symbol MAPRE3 Organism Human Gene Summary The protein encoded by this gene is a member of the RP/EB family of genes. The protein localizes to the cytoplasmic microtubule network and binds APCL a homolog of the adenomatous polyposis coli tumor suppressor gene. Gene Aliases EB3, EBF3, EBF3-S, RP3 RefSeq Accession No. NC_000002.11, NT_022184.15 UniGene ID Hs.515860 Ensembl Gene ID ENSG00000084764 Entrez Gene ID 22924 Assay Information Unique Assay ID qHsaCED0042518 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000233121, ENST00000405074, ENST00000458529, ENST00000402218 Amplicon Context Sequence GTGACCAGTGAAAATCTGAGTCGCCATGATATGCTTGCATGGGTCAACGACTCC CTGCACCTCAACTATACCAAGATAGAACAGCTTTGTTCAGGGGCA Amplicon Length (bp) 69 Chromosome Location 2:27245114-27246204 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 90 R2 0.9993 cDNA Cq 24.76 cDNA Tm (Celsius) 80 gDNA Cq 26.8 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report MAPRE3, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Human Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. -

Molecular Clock Is Involved in Predictive Circadian Adjustment of Renal Function
Molecular clock is involved in predictive circadian adjustment of renal function Annie Mercier Zubera,1,2, Gabriel Centenoa,2, Sylvain Pradervandb, Svetlana Nikolaevaa,c, Lionel Maquelina, Le´ onard Cardinauxa, Olivier Bonnya,d, and Dmitri Firsova,3 aDepartment of Pharmacology and Toxicology, University of Lausanne, 1005 Lausanne, Switzerland; bDNA Array Facility, University of Lausanne, 1015 Lausanne, Switzerland; cInstitute of Evolutionary Physiology and Biochemistry, 194223 St-Petersburg, Russia; and dService of Nephrology, Lausanne University Hospital, 1005 Lausanne, Switzerland Edited by Maurice B. Burg, National Heart, Lung, and Blood Institute, Bethesda, MD, and approved August 6, 2009 (received for review May 4, 2009) Renal excretion of water and major electrolytes exhibits a significant anticipate upcoming circadian environmental challenges (activity, circadian rhythm. This functional periodicity is believed to result, at feeding, etc). least in part, from circadian changes in secretion/reabsorption capac- The most obvious manifestation of circadian rhythmicity of renal ities of the distal nephron and collecting ducts. Here, we studied the function is a well-marked difference in the volume of urine forma- molecular mechanisms underlying circadian rhythms in the distal tion/excretion between the day and the night. The urinary excretion nephron segments, i.e., distal convoluted tubule (DCT) and connect- ϩ ϩ Ϫ Ϫ 2ϩ 2ϩ of all major solutes (Na ,K ,Cl , urea, PO4 ,Ca ,Mg )also ing tubule (CNT) and the cortical collecting duct (CCD). Temporal follows a circadian oscillating pattern. Although, renal excretion expression analysis performed on microdissected mouse DCT/CNT or rhythms are apparently synchronized with circadian rhythms of CCD revealed a marked circadian rhythmicity in the expression of a activity/feeding, they have been shown to persist over long periods large number of genes crucially involved in various homeostatic of time under experimental conditions in which external factors functions of the kidney. -

32-2748: PSMB1 Recombinant Protein Description Product Info
9853 Pacific Heights Blvd. Suite D. San Diego, CA 92121, USA Tel: 858-263-4982 Email: [email protected] 32-2748: PSMB1 Recombinant Protein Alternative HC5,PSC5,PMSB1,FLJ25321,KIAA1838,PSMB1,Proteasome subunit beta type-1,Proteasome component Name : C5,Macropain subunit C5,Multicatalytic endopeptidase complex subunit C5,Proteasome gamma chain. Description Source : Escherichia Coli. PSMB1 Human Recombinant fused to 37 amino acid His Tag at N-terminal produced in E.Coli is a single, non-glycosylated, polypeptide chain containing 250 amino acids (30-241) and having a molecular mass of 27.7 kDa. The PSMB1 is purified by proprietary chromatographic techniques. PSMB1 is part of the proteasome B-type family, also identified as the T1B family, that is a 20S core beta subunit. PSMB1 is tightly linked to the TBP (TATA-binding protein) gene in human and in mouse, and is transcribed in the opposite orientation in both species.The main function of PSMB1 is its degradation activity of unnecessary or damaged proteins by proteolysis. PSMB1 is a multicatalytic proteinase complex which is characterized by its ability to cleave peptides with arg, phe, tyr, leu, and glu adjacent to the leaving group at neutral or slightly basic ph. the proteasome has an atp-dependent proteolytic activity. Product Info Amount : 10 µg Purification : Greater than 95.0% as determined by SDS-PAGE. Content : The PSMB1 solution contains 20mM Tris-HCl pH-8, 1mM DTT and 10% glycerol. PSMB1 Recombinant Human although stable at 4°C for 30 days, should be stored desiccated Storage condition : below -20°C for periods greater than 30 days. -

The Microtubule-Associated Protein MAPRE2 Is Involved in Perineural Invasion of Pancreatic Cancer Cells
1111-1116.qxd 8/9/2009 08:18 Ì ™ÂÏ›‰·1111 INTERNATIONAL JOURNAL OF ONCOLOGY 35: 1111-1116, 2009 The microtubule-associated protein MAPRE2 is involved in perineural invasion of pancreatic cancer cells IVANE ABIATARI1*, SONJA GILLEN1*, TIAGO DeOLIVEIRA1, THERESA KLOSE1, KONG BO1, NATHALIA A. GIESE2, HELMUT FRIESS1 and JÖRG KLEEFF1,3 1Department of General Surgery, Technische Universität München, Munich; 2Department of General Surgery, University of Heidelberg, Heidelberg, Germany; 3Center of Cancer Systems Biology, Department of Medicine, Caritas St. Elizabeth's Medical Center, Tufts University School of Medicine, Boston, MA 02135-2997, USA Received May 4, 2009; Accepted July 10, 2009 DOI: 10.3892/ijo_00000426 Abstract. Perineural invasion of tumor cells is a characteristic Defining molecular mechanisms that allow pancreatic cancer feature of human pancreatic cancer. Unrevealing the molecular cells to grow along and into nerves is therefore important for mechanisms that enable cancer cells to invade and grow along the development of novel therapeutic strategies in pancreatic nerves is important for the development of novel therapeutic cancer, a disease whose incidence virtually mirrors its mortality strategies in this disease. We have previously identified tran- rate (5). We have previously identified and described in detail scriptional changes in highly nerve invasive pancreatic cancer the transcriptome signature of perineural invasion in pancreatic cells. Here we further analyzed one of the identified de- cancer by generating highly nerve invasive pancreatic cancer regulated genes, MAPRE2, a microtubule-associated protein. cells (6). One of the genes that displayed a significant up- MAPRE2 expression was significantly increased in high versus regulation in pancreatic cancer cells with a high potential for less nerve invasive pancreatic cancer cells, and changes of nerve invasion was MAPRE2. -

Identifying the Optimal Gene and Gene Set in Hepatocellular Carcinoma Based on Differential Expression and Differential Co-Expression Algorithm
1066 ONCOLOGY REPORTS 37: 1066-1074, 2017 Identifying the optimal gene and gene set in hepatocellular carcinoma based on differential expression and differential co-expression algorithm LI-YANG DONG1*, WEI-ZHONG ZHOU1*, JUN-WEI NI1, WEI XIANG1, WEN-HAO HU1, CHANG YU1 and HAI-YAN LI2 Departments of 1Invasive Technology and 2Rehabilitation, The First Affiliated Hospital of Wenzhou Medical University, Ouhai, Wenzhou, Zhejiang 325000, P.R. China Received June 23, 2016; Accepted August 10, 2016 DOI: 10.3892/or.2016.5333 * Abstract. The objective of this study was to identify the with ΔG = 18.681 and 24 HDE-HDC partitions in total. In optimal gene and gene set for hepatocellular carcinoma conclusion, we successfully investigated the optimal gene, (HCC) utilizing differential expression and differential MAPRE1, and gene set, nucleoside metabolic process, which co-expression (DEDC) algorithm. The DEDC algorithm may be potential biomarkers for targeted therapy and provide consisted of four parts: calculating differential expression significant insight for revealing the pathological mechanism (DE) by absolute t-value in t-statistics; computing differential underlying HCC. co-expression (DC) based on Z-test; determining optimal thresholds on the basis of Chi-squared (χ2) maximization and Introduction the corresponding gene was the optimal gene; and evaluating functional relevance of genes categorized into different Hepatocellular carcinoma (HCC) is the fifth most common partitions to determine the optimal gene set with highest mean cancer worldwide and the third leading cause of cancer-related * minimum functional information (FI) gain (ΔG). The optimal mortality (1), making it urgent to identify early diagnostic thresholds divided genes into four partitions, high DE and markers and therapeutic targets (2). -

King's Research Portal
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by King's Research Portal King’s Research Portal DOI: 10.1016/j.biotechadv.2017.05.004 Document Version Publisher's PDF, also known as Version of record Link to publication record in King's Research Portal Citation for published version (APA): Neville, J. J., Orlando, J., Mann, K., McCloskey, B., & Antoniou, M. N. (2017). Ubiquitous Chromatin-opening Elements (UCOEs): Applications in biomanufacturing and gene therapy. BIOTECHNOLOGY ADVANCES, 557- 564. DOI: 10.1016/j.biotechadv.2017.05.004 Citing this paper Please note that where the full-text provided on King's Research Portal is the Author Accepted Manuscript or Post-Print version this may differ from the final Published version. If citing, it is advised that you check and use the publisher's definitive version for pagination, volume/issue, and date of publication details. And where the final published version is provided on the Research Portal, if citing you are again advised to check the publisher's website for any subsequent corrections. General rights Copyright and moral rights for the publications made accessible in the Research Portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognize and abide by the legal requirements associated with these rights. •Users may download and print one copy of any publication from the Research Portal for the purpose of private study or research. •You may not further distribute the material or use it for any profit-making activity or commercial gain •You may freely distribute the URL identifying the publication in the Research Portal Take down policy If you believe that this document breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Evidence for Differential Alternative Splicing in Blood of Young Boys With
Stamova et al. Molecular Autism 2013, 4:30 http://www.molecularautism.com/content/4/1/30 RESEARCH Open Access Evidence for differential alternative splicing in blood of young boys with autism spectrum disorders Boryana S Stamova1,2,5*, Yingfang Tian1,2,4, Christine W Nordahl1,3, Mark D Shen1,3, Sally Rogers1,3, David G Amaral1,3 and Frank R Sharp1,2 Abstract Background: Since RNA expression differences have been reported in autism spectrum disorder (ASD) for blood and brain, and differential alternative splicing (DAS) has been reported in ASD brains, we determined if there was DAS in blood mRNA of ASD subjects compared to typically developing (TD) controls, as well as in ASD subgroups related to cerebral volume. Methods: RNA from blood was processed on whole genome exon arrays for 2-4–year-old ASD and TD boys. An ANCOVA with age and batch as covariates was used to predict DAS for ALL ASD (n=30), ASD with normal total cerebral volumes (NTCV), and ASD with large total cerebral volumes (LTCV) compared to TD controls (n=20). Results: A total of 53 genes were predicted to have DAS for ALL ASD versus TD, 169 genes for ASD_NTCV versus TD, 1 gene for ASD_LTCV versus TD, and 27 genes for ASD_LTCV versus ASD_NTCV. These differences were significant at P <0.05 after false discovery rate corrections for multiple comparisons (FDR <5% false positives). A number of the genes predicted to have DAS in ASD are known to regulate DAS (SFPQ, SRPK1, SRSF11, SRSF2IP, FUS, LSM14A). In addition, a number of genes with predicted DAS are involved in pathways implicated in previous ASD studies, such as ROS monocyte/macrophage, Natural Killer Cell, mTOR, and NGF signaling. -

Epigenome-Wide Exploratory Study of Monozygotic Twins Suggests Differentially Methylated Regions to Associate with Hand Grip Strength
Biogerontology (2019) 20:627–647 https://doi.org/10.1007/s10522-019-09818-1 (0123456789().,-volV)( 0123456789().,-volV) RESEARCH ARTICLE Epigenome-wide exploratory study of monozygotic twins suggests differentially methylated regions to associate with hand grip strength Mette Soerensen . Weilong Li . Birgit Debrabant . Marianne Nygaard . Jonas Mengel-From . Morten Frost . Kaare Christensen . Lene Christiansen . Qihua Tan Received: 15 April 2019 / Accepted: 24 June 2019 / Published online: 28 June 2019 Ó The Author(s) 2019 Abstract Hand grip strength is a measure of mus- significant CpG sites or pathways were found, how- cular strength and is used to study age-related loss of ever two of the suggestive top CpG sites were mapped physical capacity. In order to explore the biological to the COL6A1 and CACNA1B genes, known to be mechanisms that influence hand grip strength varia- related to muscular dysfunction. By investigating tion, an epigenome-wide association study (EWAS) of genomic regions using the comb-p algorithm, several hand grip strength in 672 middle-aged and elderly differentially methylated regions in regulatory monozygotic twins (age 55–90 years) was performed, domains were identified as significantly associated to using both individual and twin pair level analyses, the hand grip strength, and pathway analyses of these latter controlling the influence of genetic variation. regions revealed significant pathways related to the Moreover, as measurements of hand grip strength immune system, autoimmune disorders, including performed over 8 years were available in the elderly diabetes type 1 and viral myocarditis, as well as twins (age 73–90 at intake), a longitudinal EWAS was negative regulation of cell differentiation. -

Cellular and Molecular Signatures in the Disease Tissue of Early
Cellular and Molecular Signatures in the Disease Tissue of Early Rheumatoid Arthritis Stratify Clinical Response to csDMARD-Therapy and Predict Radiographic Progression Frances Humby1,* Myles Lewis1,* Nandhini Ramamoorthi2, Jason Hackney3, Michael Barnes1, Michele Bombardieri1, Francesca Setiadi2, Stephen Kelly1, Fabiola Bene1, Maria di Cicco1, Sudeh Riahi1, Vidalba Rocher-Ros1, Nora Ng1, Ilias Lazorou1, Rebecca E. Hands1, Desiree van der Heijde4, Robert Landewé5, Annette van der Helm-van Mil4, Alberto Cauli6, Iain B. McInnes7, Christopher D. Buckley8, Ernest Choy9, Peter Taylor10, Michael J. Townsend2 & Costantino Pitzalis1 1Centre for Experimental Medicine and Rheumatology, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK. Departments of 2Biomarker Discovery OMNI, 3Bioinformatics and Computational Biology, Genentech Research and Early Development, South San Francisco, California 94080 USA 4Department of Rheumatology, Leiden University Medical Center, The Netherlands 5Department of Clinical Immunology & Rheumatology, Amsterdam Rheumatology & Immunology Center, Amsterdam, The Netherlands 6Rheumatology Unit, Department of Medical Sciences, Policlinico of the University of Cagliari, Cagliari, Italy 7Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8TA, UK 8Rheumatology Research Group, Institute of Inflammation and Ageing (IIA), University of Birmingham, Birmingham B15 2WB, UK 9Institute of -

The MAP4K4-STRIPAK Complex Promotes Growth and Tissue Invasion In
bioRxiv preprint doi: https://doi.org/10.1101/2021.05.07.442906; this version posted May 8, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 The MAP4K4-STRIPAK complex promotes growth and tissue invasion in 2 medulloblastoma 3 Jessica Migliavacca1, Buket Züllig1, Charles Capdeville1, Michael Grotzer2 and Martin Baumgartner1,* 4 1 Division of Oncology, Children’s Research Center, University Children’s Hospital Zürich, Zürich, 5 Switzerland 6 2 Division of Oncology, University Children’s Hospital Zürich, Zürich, Switzerland 7 8 *e-mail: [email protected] 9 10 Abstract 11 Proliferation and motility are mutually exclusive biological processes associated with cancer that depend 12 on precise control of upstream signaling pathways with overlapping functionalities. We find that STRN3 13 and STRN4 scaffold subunits of the STRIPAK complex interact with MAP4K4 for pathway regulation in 14 medulloblastoma. Disruption of the MAP4K4-STRIPAK complex impairs growth factor-induced 15 migration and tissue invasion and stalls YAP/TAZ target gene expression and oncogenic growth. The 16 migration promoting functions of the MAP4K4-STRIPAK complex involve the activation of novel PKCs 17 and the phosphorylation of the membrane targeting S157 residue of VASP through MAP4K4. The anti- 18 proliferative effect of complex disruption is associated with reduced YAP/TAZ target gene expression 19 and results in repressed tumor growth in the brain tissue. This dichotomous functionality of the STRIPAK 20 complex in migration and proliferation control acts through MAP4K4 regulation in tumor cells and 21 provides relevant mechanistic insights into novel tumorigenic functions of the STRIPAK complex in 22 medulloblastoma.