Hepadnaviridae
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Hepatitis Virus in Long-Fingered Bats, Myanmar
DISPATCHES Myanmar; the counties are adjacent to Yunnan Province, Hepatitis Virus People’s Republic of China. The bats covered 6 species: Miniopterus fuliginosus (n = 640), Hipposideros armiger in Long-Fingered (n = 8), Rhinolophus ferrumequinum (n = 176), Myotis chi- nensis (n = 11), Megaderma lyra (n = 6), and Hipposideros Bats, Myanmar fulvus (n = 12). All bat tissue samples were subjected to vi- Biao He,1 Quanshui Fan,1 Fanli Yang, ral metagenomic analysis (unpublished data). The sampling Tingsong Hu, Wei Qiu, Ye Feng, Zuosheng Li, of bats for this study was approved by the Administrative Yingying Li, Fuqiang Zhang, Huancheng Guo, Committee on Animal Welfare of the Institute of Military Xiaohuan Zou, and Changchun Tu Veterinary, Academy of Military Medical Sciences, China. We used PCR to further study the prevalence of or- During an analysis of the virome of bats from Myanmar, thohepadnavirus in the 6 bat species; the condition of the a large number of reads were annotated to orthohepadnavi- samples made serologic assay and pathology impracticable. ruses. We present the full genome sequence and a morpho- Viral DNA was extracted from liver tissue of each of the logical analysis of an orthohepadnavirus circulating in bats. 853 bats by using the QIAamp DNA Mini Kit (QIAGEN, This virus is substantially different from currently known Hilden, Germany). To detect virus in the samples, we con- members of the genus Orthohepadnavirus and represents ducted PCR by using the TaKaRa PCR Kit (TaKaRa, Da- a new species. lian, China) with a pair of degenerate pan-orthohepadnavi- rus primers (sequences available upon request). The PCR he family Hepadnaviridae comprises 2 genera (Ortho- reaction was as follows: 45 cycles of denaturation at 94°C Thepadnavirus and Avihepadnavirus), and viruses clas- for 30 s, annealing at 54°C for 30 s, extension at 72°C for sified within these genera have a narrow host range. -

Intracellular Trafficking of HBV Particles
cells Review Intracellular Trafficking of HBV Particles Bingfu Jiang 1 and Eberhard Hildt 1,2,* 1 Department of Virology, Paul-Ehrlich-Institut, D-63225 Langen, Germany; [email protected] 2 German Center for Infection Research (DZIF), TTU Hepatitis, Marburg-Gießen-Langen, D63225 Langen, Germany * Correspondence: [email protected]; Tel.: +49-61-0377-2140 Received: 12 August 2020; Accepted: 2 September 2020; Published: 2 September 2020 Abstract: The human hepatitis B virus (HBV), that is causative for more than 240 million cases of chronic liver inflammation (hepatitis), is an enveloped virus with a partially double-stranded DNA genome. After virion uptake by receptor-mediated endocytosis, the viral nucleocapsid is transported towards the nuclear pore complex. In the nuclear basket, the nucleocapsid disassembles. The viral genome that is covalently linked to the viral polymerase, which harbors a bipartite NLS, is imported into the nucleus. Here, the partially double-stranded DNA genome is converted in a minichromosome-like structure, the covalently closed circular DNA (cccDNA). The DNA virus HBV replicates via a pregenomic RNA (pgRNA)-intermediate that is reverse transcribed into DNA. HBV-infected cells release apart from the infectious viral parrticle two forms of non-infectious subviral particles (spheres and filaments), which are assembled by the surface proteins but lack any capsid and nucleic acid. In addition, naked capsids are released by HBV replicating cells. Infectious viral particles and filaments are released via multivesicular bodies; spheres are secreted by the classic constitutive secretory pathway. The release of naked capsids is still not fully understood, autophagosomal processes are discussed. This review describes intracellular trafficking pathways involved in virus entry, morphogenesis and release of (sub)viral particles. -

Virus Particle Structures
Virus Particle Structures Virus Particle Structures Palmenberg, A.C. and Sgro, J.-Y. COLOR PLATE LEGENDS These color plates depict the relative sizes and comparative virion structures of multiple types of viruses. The renderings are based on data from published atomic coordinates as determined by X-ray crystallography. The international online repository for 3D coordinates is the Protein Databank (www.rcsb.org/pdb/), maintained by the Research Collaboratory for Structural Bioinformatics (RCSB). The VIPER web site (mmtsb.scripps.edu/viper), maintains a parallel collection of PDB coordinates for icosahedral viruses and additionally offers a version of each data file permuted into the same relative 3D orientation (Reddy, V., Natarajan, P., Okerberg, B., Li, K., Damodaran, K., Morton, R., Brooks, C. and Johnson, J. (2001). J. Virol., 75, 11943-11947). VIPER also contains an excellent repository of instructional materials pertaining to icosahedral symmetry and viral structures. All images presented here, except for the filamentous viruses, used the standard VIPER orientation along the icosahedral 2-fold axis. With the exception of Plate 3 as described below, these images were generated from their atomic coordinates using a novel radial depth-cue colorization technique and the program Rasmol (Sayle, R.A., Milner-White, E.J. (1995). RASMOL: biomolecular graphics for all. Trends Biochem Sci., 20, 374-376). First, the Temperature Factor column for every atom in a PDB coordinate file was edited to record a measure of the radial distance from the virion center. The files were rendered using the Rasmol spacefill menu, with specular and shadow options according to the Van de Waals radius of each atom. -

Discovery of a Highly Divergent Hepadnavirus in Shrews from China
Virology 531 (2019) 162–170 Contents lists available at ScienceDirect Virology journal homepage: www.elsevier.com/locate/virology Discovery of a highly divergent hepadnavirus in shrews from China T Fang-Yuan Niea,b,1, Jun-Hua Tianc,1, Xian-Dan Lind,1, Bin Yuc, Jian-Guang Xinge, Jian-Hai Caof, ⁎ ⁎⁎ Edward C. Holmesg,h, Runlin Z. Maa, , Yong-Zhen Zhangb,h, a State Key Laboratory of Molecular Developmental Biology, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China b State Key Laboratory for Infectious Disease Prevention and Control; Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases; Department of Zoonoses, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Changping, Beijing, China c Wuhan Center for Disease Control and Prevention, Wuhan, China d Wenzhou Center for Disease Control and Prevention, Wenzhou, Zhejiang Province, China e Wencheng Center for Disease Control and Prevention, Wencheng, Zhejiang Province, China f Longwan Center for Disease Control and Prevention, Longwan District, Wenzhou, Zhejiang Province, China g Marie Bashir Institute for Infectious Diseases and Biosecurity, Charles Perkins Centre, School of Life and Environmental Sciences and Sydney Medical School, The University of Sydney, Sydney, NSW 2006, Australia h Shanghai Public Health Clinical Center & Institute of Biomedical Sciences, Fudan University, Shanghai, China ARTICLE INFO ABSTRACT Keywords: Limited sampling means that relatively little is known about the diversity and evolutionary history of mam- Hepadnaviruses malian members of the Hepadnaviridae (genus Orthohepadnavirus). An important case in point are shrews, the Shrews fourth largest group of mammals, but for which there is limited knowledge on the role they play in viral evo- Phylogeny lution and emergence. -

And Giant Guitarfish (Rhynchobatus Djiddensis)
VIRAL DISCOVERY IN BLUEGILL SUNFISH (LEPOMIS MACROCHIRUS) AND GIANT GUITARFISH (RHYNCHOBATUS DJIDDENSIS) BY HISTOPATHOLOGY EVALUATION, METAGENOMIC ANALYSIS AND NEXT GENERATION SEQUENCING by JENNIFER ANNE DILL (Under the Direction of Alvin Camus) ABSTRACT The rapid growth of aquaculture production and international trade in live fish has led to the emergence of many new diseases. The introduction of novel disease agents can result in significant economic losses, as well as threats to vulnerable wild fish populations. Losses are often exacerbated by a lack of agent identification, delay in the development of diagnostic tools and poor knowledge of host range and susceptibility. Examples in bluegill sunfish (Lepomis macrochirus) and the giant guitarfish (Rhynchobatus djiddensis) will be discussed here. Bluegill are popular freshwater game fish, native to eastern North America, living in shallow lakes, ponds, and slow moving waterways. Bluegill experiencing epizootics of proliferative lip and skin lesions, characterized by epidermal hyperplasia, papillomas, and rarely squamous cell carcinoma, were investigated in two isolated poopulations. Next generation genomic sequencing revealed partial DNA sequences of an endogenous retrovirus and the entire circular genome of a novel hepadnavirus. Giant Guitarfish, a rajiform elasmobranch listed as ‘vulnerable’ on the IUCN Red List, are found in the tropical Western Indian Ocean. Proliferative skin lesions were observed on the ventrum and caudal fin of a juvenile male quarantined at a public aquarium following international shipment. Histologically, lesions consisted of papillomatous epidermal hyperplasia with myriad large, amphophilic, intranuclear inclusions. Deep sequencing and metagenomic analysis produced the complete genomes of two novel DNA viruses, a typical polyomavirus and a second unclassified virus with a 20 kb genome tentatively named Colossomavirus. -

Methylation Profile of Hepatitis B Virus Is Not Influenced by Interferon Α in Human Liver Cancer Cells
MOLECULAR MEDICINE REPORTS 24: 715, 2021 Methylation profile of hepatitis B virus is not influenced by interferon α in human liver cancer cells IN YOUNG MOON1 and JIN‑WOOK KIM1,2 1Department of Medicine, Seoul National University Bundang Hospital, Seongnam, Gyeonggi 13620; 2Department of Internal Medicine, Seoul National University College of Medicine, Seoul 03080, Republic of Korea Received December 20, 2020; Accepted July 12, 2021 DOI: 10.3892/mmr.2021.12354 Abstract. Interferon (IFN) α is used for the treatment of Introduction chronic hepatitis B virus (HBV) infection, but the molecular mechanisms underlying its antiviral effect have not been fully Hepatitis B virus (HBV) is one of the commonest causes elucidated. Epigenetic modifications regulate the transcrip‑ of chronic hepatitis worldwide (1). The chronicity of HBV tional activity of covalently closed circular DNA (cccDNA) in infection is related to the fact that HBV produces very cells with chronic HBV infection. IFN‑α has been shown to stable viral genome in the host cells (1). The HBV genome modify cccDNA‑bound histones, but it is not known whether is partially double‑stranded relaxed circular DNA (rcDNA), the anti‑HBV effect of IFN‑α involves methylation of cccDNA. which is converted to complete double‑stranded covalently The present study aimed to determine whether IFN‑α induced closed circular DNA (cccDNA) in the nucleus of infected methylation of HBV cccDNA in a cell‑based model in which hepatocytes (2). HBV cccDNA remains the main hurdle in the HepG2 cells were directly infected with wild‑type HBV eradication of infected HBV as current antiviral agents cannot virions. -

Quantification and Epigenetic Evaluation of the Residual Pool Of
www.nature.com/scientificreports OPEN Quantifcation and epigenetic evaluation of the residual pool of hepatitis B covalently closed circular DNA in long‑term nucleoside analogue‑treated patients Fanny Lebossé1,2,3, Aurore Inchauspé1,2, Maëlle Locatelli1,2, Clothilde Miaglia1,2,3, Audrey Diederichs1,2, Judith Fresquet1,2, Fleur Chapus1,2, Kamal Hamed4, Barbara Testoni1,2* & Fabien Zoulim1,2,3* Hepatitis B virus (HBV) covalently closed circular (ccc)DNA is the key genomic form responsible for viral persistence and virological relapse after treatment withdrawal. The assessment of residual intrahepatic cccDNA levels and activity after long‑term nucleos(t)ide analogues therapy still represents a technical challenge. Quantitative (q)PCR, rolling circle amplifcation (RCA) and droplet digital (dd)PCR assays were used to quantify residual intrahepatic cccDNA in liver biopsies from 56 chronically HBV infected patients after 3 to 5 years of telbivudine treatment. Activity of residual cccDNA was evaluated by quantifying 3.5 kB HBV RNA (preC/pgRNA) and by assessing cccDNA‑associated histone tails post‑transcriptional modifcations (PTMs) by micro‑chromatin immunoprecipitation. Long‑term telbivudine treatment resulted in serum HBV DNA suppression, with most of the patients reaching undetectable levels. Despite 38 out of 56 patients had undetectable cccDNA when assessed by qPCR, RCA and ddPCR assays detected cccDNA in all‑but‑one negative samples. Low preC/pgRNA level in telbivudine‑treated samples was associated with enrichment for cccDNA histone PTMs related to repressed transcription. No diference in cccDNA levels was found according to serum viral markers evolution. This panel of cccDNA evaluation techniques should provide an added value for the new proof‑of‑concept clinical trials aiming at a functional cure of chronic hepatitis B. -
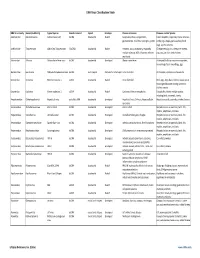
Virus Classification Tables V2.Vd.Xlsx
DNA Virus Classification Table DNA Virus Family Genera (Subfamily) Typical Species Genetic material Capsid Envelope Disease in Humans Diseases in other Species Adenoviridae Mastadenovirus Adenoviruses 1‐47 dsDNA Icosahedral Naked Respiratory illness; conjunctivitis, Canine hepatitis, respiratory illness in horses, gastroenteritis, tonsillitis, meningitis, cystitis cattle, pigs, sheep, goats, sea lions, birds dogs, squirrel enteritis Anelloviridae Torqueviruses Alpha‐Zeta Torqueviruses (‐)ssDNA Icosahedral Naked Hepatitis, lupus, pulmonary, myopathy, Chimpanzee, pig, cow, sheep, tree shrews, multiple sclerosis; 90% of humans infected pigs, cats, sea lions and chickens worldwide Asfarviridae Asfivirus African Swine fever virus dsDNA Icosahedral Enveloped African swine fever Arthropod (tick) transmission or ingestion; hemorrhagic fever in warthogs, pigs Baculoviridae Baculovirus Alpha‐Gamma Baculoviruses dsDNA Stick shaped Occluded or Enveloped none identified Arthropods, Lepidoptera, crustaceans Circoviridae Circovirus Porcine circovirus 1 ssDNA Icosahedral Naked none identified Birds, pigs, dogs; bats; rodents; causes post‐ weaning multisystem wasting syndrome, chicken anemia Circoviridae Cyclovirus Human cyclovirus 1 ssDNA Icosahedral Naked Cyclovirus Vietnam encephalitis Encephalitis; infects multiple species including birds, mammals, insects Hepadnaviridae Orthohepadnavirus Hepatitis B virus partially ssDNA Icosahedral Enveloped Hepatitis B virus; Cirrhosis, Hepatocellular Hepatitis in ducks, squirrels, primates, herons carcinoma Herpesviridae -

Summary of Antimicrobial Activity
SUMMARY OF ANTIMICROBIAL ACTIVITY 3x RENEGADE DAILY ONE-STEP DISINFECTANT Description 3x RENEGADE DAILY Disinfectant & Detergent is a broad spectrum, hard surface disinfectant. When used as directed, this product will deliver effective biocidal action against bacteria, fungi, and viruses. This formulation is a blend of a premium active ingredients and inerts: surfactants, chelates, and water. Biocidal performance is attained when this product is properly diluted at 1/2 oz. per gallon or 1:256 (1 oz. per gallon or 1:128 for Norovirus). 3x RENEGADE DAILY can be used to disinfect a wide variety of hard surfaces such as floors, walls, toilets, sinks, and countertops in hospitals, households, and institutions. Regulatory Summary Physical Properties EPA Registration No. 6836-349- pH of Concentrate 12.0 – 13.5 Flash Point (PMCC) >200 F 12120 USDA Authorization None Specific Gravity @ 0.98 – 1.05 g/mL % Quat (mol. wt.342.0) 22.24 25°C California Status Pounds per gallon @ 8.42 – 8.51 % Volatile 93.5-94.5 25°C Canadian PCP# None Canadian Din # None Summary of Antimicrobial Test Results 3x RENEGADE DAILY is a "One-Step" Hospital Disinfectant, Virucide, Fungicide, Mildewstat, Sanitizer and Cleaner. Listed in the following pages is a summary of Antimicrobial Claims and a review of test results. Claim: Contact time: Organic Soil: Water Conditions: Disinfectant Varies 5% 250ppm as CaCO3 Test Method: AOAC Germicidal Spray Test Organism Contact Dilution Time (Min) 868 ppm (1/2oz. per Acinetobacter baumannii 3 Gal) Bordetella bronchiseptica 3 868 ppm Bordetella pertussis 3 868 ppm Campylobacter jejuni 3 868 ppm Enterobacter aerogenes 3 1736 PPM (1 oz per Gal) Enterococcus faecalis 3 868 ppm Enterococcus faecalis - Vancomycin resistant [VRE] 3 868 ppm Escherichia coli 3 868 ppm Escherichia coli [O157:H7] 3 868 ppm Escherichia coli ESBL – Extended spectrum beta- 868 ppm 10 lactamase containing E. -

In Vitro Selection and Characterization of Single Stranded DNA Aptamers Inhibiting the Hepatitis B Virus Capsid-Envelope Interaction
Aus dem Veterinärwissenschaftlichen Department der Tierärztlichen Fakultät der Ludwig-Maximilians-Universität München Arbeit angefertigt unter der Leitung von: Univ.-Prof. Dr. Gerd Sutter Angefertigt im Institut für Virologie des Helmholtz Zentrum München (apl.-Prof. Dr. Volker Bruss) In vitro Selection and Characterization of single stranded DNA Aptamers Inhibiting the Hepatitis B Virus Capsid-Envelope Interaction Inaugural-Dissertation zur Erlangung der tiermedizinischen Doktorwürde der Tierärztlichen Fakultät der Ludwig-Maximilians-Universität München von Ahmed El-Sayed Abd El-Halem Orabi aus Sharkia/Ägypten München 2013 Gedruckt mit der Genehmigung der Tierärztlichen Fakultät der Ludwig-Maximilians-Universität München Dekan: Univ.-Prof. Dr. Joachim Braun Berichterstatter: Univ.-Prof. Dr. Gerd Sutter Korreferent: Univ.-Prof. Dr. Bernd Kaspers Tag der Promotion: 20. Juli 2013 My Family Contents Contents 1 INTRODUCTION ................................................................................................... 1 2 REVIEW OF THE LITERATURE ....................................................................... 2 2.1 HEPATITIS B VIRUS (HBV) .............................................................................................. 2 2.1.1 HISTORY AND TAXONOMY .......................................................................................................... 2 2.1.2 EPIDEMIOLOGY AND PATHOGENESIS .......................................................................................... 3 2.1.3 VIRION STRUCTURE .................................................................................................................... -

Inhibition of Jcpyv Infection Mediated by Targeted Viral Genome Editing
www.nature.com/scientificreports OPEN Inhibition of JCPyV infection mediated by targeted viral genome editing using CRISPR/Cas9 Received: 31 May 2016 Yi-ying Chou1, Annabel Krupp2, Campbell Kaynor2, Raphaël Gaudin1,†, Minghe Ma1, Accepted: 21 October 2016 Ellen Cahir-McFarland2 & Tom Kirchhausen1,3 Published: 14 November 2016 Progressive multifocal leukoencephalopathy (PML) is a debilitating disease resulting from infection of oligodendrocytes by the JC polyomavirus (JCPyV). Currently, there is no anti-viral therapeutic available against JCPyV infection. The clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR- associated protein 9 (Cas9) system (CRISPR/Cas9) is a genome editing tool capable of introducing sequence specific breaks in double stranded DNA. Here we show that the CRISPR/Cas9 system can restrict the JCPyV life cycle in cultured cells. We utilized CRISPR/Cas9 to target the noncoding control region and the late gene open reading frame of the JCPyV genome. We found significant inhibition of virus replication and viral protein expression in cells recipient of Cas9 together with JCPyV-specific single-guide RNA delivered prior to or after JCPyV infection. The JC polyomavirus (JCPyV), a member of the Polyomaviridae family, is the causative agent of a rare and debili- tating demyelinating disease termed progressive multifocal leukoencephalopathy (PML) caused by infection and destruction of oligodendrocytes in the central nervous system (CNS)1,2. PML can develop in the context of immu- nodeficiency and treatment with immunomodulatory drugs3. Indeed, it is one of the most common CNS-related diseases in AIDS, affecting 5% of the HIV-1 positive patients. The incidence of PML has also risen to 0.2–0.4% in patients receiving immunomodulatory therapies4,5. -

Viral Epigenomes in Human Tumorigenesis
Oncogene (2010) 29, 1405–1420 & 2010 Macmillan Publishers Limited All rights reserved 0950-9232/10 $32.00 www.nature.com/onc REVIEW Viral epigenomes in human tumorigenesis AF Fernandez1 and M Esteller1,2 1Cancer Epigenetics and Biology Program (PEBC), Bellvitge Biomedical Research Institute (IDIBELL), Barcelona, Catalonia, Spain and 2Institucio Catalana de Recerca i Estudis Avanc¸ats (ICREA), Barcelona, Catalonia, Spain Viruses are associated with 15–20% of human cancers is altered in cancer (Fraga and Esteller, 2005; Chuang worldwide. In the last century, many studies were directed and Jones, 2007; Lujambio et al., 2007). towards elucidating the molecular mechanisms and genetic DNA methylation mainly occurs on cytosines that alterations by which viruses cause cancer. The importance precede guanines to yield 5-methylcytosine; these of epigenetics in the regulation of gene expression has dinucleotide sites are usually referred to as CpGs prompted the investigation of virus and host interactions (Herman and Baylin, 2003). CpGs are asymmetrically not only at the genetic level but also at the epigenetic level. distributed into CpG-poor regions and dense regions In this study, we summarize the published epigenetic called ‘CpG islands’, which are located in the promoter information relating to the genomes of viruses directly or regions of approximately half of all genes. These CpG indirectly associated with the establishment of tumori- islands are usually unmethylated in normal cells, with genic processes. We also review aspects such as viral the exceptions listed below, whereas the sporadic CpG replication and latency associated with epigenetic changes sites in the rest of the genome are generally methylated and summarize what is known about epigenetic alterations (Jones and Takai, 2001).