Cycad (Cycadales) Chromosome Numbers Are Not Correlated with Genome Size
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

International Organisation of Palaeobotany IOP NEWSLETTER
INTERNATIONAL UNION OF BIOLOGIC A L S C IENC ES S ECTION FOR P A L A EOBOTANY International Organisation of Palaeobotany IOP NEWSLETTER 110 August 2016 CONTENTS FROM THE SECRETARY/TREASURER IPC XIV/IOPC X 2016 IOPC 2020 IOP MEMBERSHIP IOP EXECUTIVE COMMITTEE ELECTIONS IOP WEBMASTER POSITION WHAT HAPPENED TO THE OUPH COLLECTIONS? THE PALAEOBOTANY OF ITALY UPCOMING MEETINGS CALL FOR NEWS and NOTES The views expressed in the newsletter are those of its correspondents, and do not necessarily reflect the policy of IOP. Please send us your contributions for the next edition of our newsletter (June 2016) by M ay 30th, 2016. President: Johanna Eder-Kovar (G ermany) Vice Presidents: Bob Spicer (Great Britain), Harufumi Nishida (Japan), M ihai Popa (Romania) M embers at Large: Jun W ang (China), Hans Kerp (Germany), Alexej Herman (Russia) Secretary/Treasurer/Newsletter editor: M ike Dunn (USA) Conference/Congress Chair: Francisco de Assis Ribeiro dos Santos IOP Logo: The evolution of plant architecture (© by A. R. Hemsley) I OP 110 2 August 2016 FROM THE In addition, please send any issues that you think need to be addressed at the Business SECRETARY/TREASURER meeting. I will add those to the Agenda. Dear IOP Members, Respectfully, Mike I am happy to report, that IOP seems to be on track and ready for a new Executive Council to take over. The elections are IPC XIV/IOPC X 2016 progressing nicely and I will report the results in the September/October Newsletter. The one area that is still problematic is the webmaster position. We really to talk amongst ourselves, and find someone who is willing and able to do the job. -

Chromosome Numbers in Gymnosperms - an Update
Rastogi and Ohri . Silvae Genetica (2020) 69, 13 - 19 13 Chromosome Numbers in Gymnosperms - An Update Shubhi Rastogi and Deepak Ohri Amity Institute of Biotechnology, Research Cell, Amity University Uttar Pradesh, Lucknow Campus, Malhaur (Near Railway Station), P.O. Chinhat, Luc know-226028 (U.P.) * Corresponding author: Deepak Ohri, E mail: [email protected], [email protected] Abstract still some controversy with regard to a monophyletic or para- phyletic origin of the gymnosperms (Hill 2005). Recently they The present report is based on a cytological data base on 614 have been classified into four subclasses Cycadidae, Ginkgoi- (56.0 %) of the total 1104 recognized species and 82 (90.0 %) of dae, Gnetidae and Pinidae under the class Equisetopsida the 88 recognized genera of gymnosperms. Family Cycada- (Chase and Reveal 2009) comprising 12 families and 83 genera ceae and many genera of Zamiaceae show intrageneric unifor- (Christenhusz et al. 2011) and 88 genera with 1104 recognized mity of somatic numbers, the genus Zamia is represented by a species according to the Plant List (www.theplantlist.org). The range of number from 2n=16-28. Ginkgo, Welwitschia and Gen- validity of accepted name of each taxa and the total number of tum show 2n=24, 2n=42, and 2n=44 respectively. Ephedra species in each genus has been checked from the Plant List shows a range of polyploidy from 2x-8x based on n=7. The (www.theplantlist.org). The chromosome numbers of 688 taxa family Pinaceae as a whole shows 2n=24except for Pseudolarix arranged according to the recent classification (Christenhusz and Pseudotsuga with 2n=44 and 2n=26 respectively. -

A New Microsporangiate Organ from the Lower Carboniferous of the Novgorod Region, Russia O
ISSN 0031-0301, Paleontological Journal, 2009, Vol. 43, No. 10, pp. 1316–1329. © Pleiades Publishing, Ltd., 2009. A New Microsporangiate Organ from the Lower Carboniferous of the Novgorod Region, Russia O. A. Orlovaa, N. R. Meyer-Melikian†,a, and N. E. Zavialovab a Moscow State University (MGU), Leninskie gory 1, Moscow, 119991 Russia b Borissiak Paleontological Institute of the Russian Academy of Sciences, ul. Profsoyuznaya 123, Moscow, 117997 Russia e-mail: [email protected] Received February 5, 2008 Abstract—A new species of the genus Telangiopsis, T. nonnae O. Orlova et Zavialova, was described on the basis of a microsporangiate organ from the Lower Carboniferous deposits of the Novgorod Region. The mor- phology of branching fertile axes, synangia, and sporangia was thoroughly studied. The three-dimensional sys- tem of fertile axes branches monopodially; ultimate axes bear numerous connivent bunches of synangia, which consist of three to six basally fused elongated ovate sporangia. The morphology and ultrastructure of prepollen grains were studied, which were extracted from the rock matrix surrounding the sporangia. The two-layered exine includes a well-developed endexine and an alveolate ectexine, with one–three rows of large thin-walled alveolae. The new species was compared with other Early Carboniferous microsporangiate organs. Key words: Early Carboniferous, Novgorod Region, fertile axis, synangia, Lyginopteridales, trilete prepollen, exine ultrastructure. DOI: 10.1134/S003103010910013X INTRODUCTION synangia and numerous casts and imprints of detached synangia were found in yellow ferruginous sandstone. During the three last decades, the interest of bota- In addition to the fertile axes, sterile remains of Lygi- nists dealing with fossil and modern plants to Carbon- nopteridales, Medullosales, and Calamopytiales were iferous synangiate pollen organs has considerably found (Orlova and Snigirevskii, 2003, 2004). -

Dr. Sahanaj Jamil Associate Professor of Botany M.L.S.M. College, Darbhanga
Subject BOTANY Paper No V Paper Code BOT521 Topic Taxonomy and Diversity of Seed Plant: Gymnosperms & Angiosperms Dr. Sahanaj Jamil Associate Professor of Botany M.L.S.M. College, Darbhanga BOTANY PG SEMESTER – II, PAPER –V BOT521: Taxonomy and Diversity of seed plants UNIT- I BOTANY PG SEMESTER – II, PAPER –V BOT521: Taxonomy and Diversity of seed plants Classification of Gymnosperms. # Robert Brown (1827) for the first time recognized Gymnosperm as a group distinct from angiosperm due to the presence of naked ovules. BENTHAM and HOOKSER (1862-1883) consider them equivalent to dicotyledons and monocotyledons and placed between these two groups of angiosperm. They recognized three classes of gymnosperm, Cyacadaceae, coniferac and gnetaceae. Later ENGLER (1889) created a group Gnikgoales to accommodate the genus giankgo. Van Tieghem (1898) treated Gymnosperm as one of the two subdivision of spermatophyte. To accommodate the fossil members three more classes- Pteridospermae, Cordaitales, and Bennettitales where created. Coulter and chamberlain (1919), Engler and Prantl (1926), Rendle (1926) and other considered Gymnosperm as a division of spermatophyta, Phanerogamia or Embryoptyta and they further divided them into seven orders: - i) Cycadofilicales ii) Cycadales iii) Bennettitales iv) Ginkgoales v) Coniferales vi) Corditales vii) Gnetales On the basis of wood structure steward (1919) divided Gymnosperm into two classes: - i) Manoxylic ii) Pycnoxylic The various classification of Gymnosperm proposed by various workers are as follows: - i) Sahni (1920): - He recognized two sub-divison in gymnosperm: - a) Phylospermae b) Stachyospermae BOTANY PG SEMESTER – II, PAPER –V BOT521: Taxonomy and Diversity of seed plants ii) Classification proposed by chamber lain (1934): - He divided Gymnosperm into two divisions: - a) Cycadophyta b) Coniterophyta iii) Classification proposed by Tippo (1942):- He considered Gymnosperm as a class of the sub- phylum pteropsida and divided them into two sub classes:- a) Cycadophyta b) Coniferophyta iv) D. -

Organization of a Dispersed Repeated DNA Element in the Zamia Genome
BIOLOGIA PLANTARUM 53 (1): 28-36, 2009 Organization of a dispersed repeated DNA element in the Zamia genome D. CAFASSO1*, S. COZZOLINO1, N.J. VEREECKEN2, P. DE LUCA3 and G. CHINALI4 Dipartimento di Biologia Strutturale e Funzionale, Complesso Universitario Monte S. Angelo, Università degli Studi di Napoli Federico II, Via Cinthia, I-80126 Napoli, Italy1 Behavioural and Evolutionary Ecology, Free University of Brussels, Av. F.D. Roosevelt 50, B-1050 Brussels, Belgium2 Dipartimento delle Scienze Biologiche, Università degli Studi di Napoli Federico II, Via Foria 223, I-80139 Napoli, Italy3 Dipartimento di Medicina Clinica e Sperimentale, Facoltà di Medicina e Chirurgia, Università di Napoli Federico II. Via Pansini 5, I-80131 Napoli, Italy4 Abstract Occurrence and genomic organization of dispersed elements containing ZpS1 satellite repeats have been investigated in a wide representation of species of the old plant genus Zamia (Zamiaceae, Cycadales). In Z. paucijuga, the ZpS1 repeat is organized as long satellite DNA arrays and as short arrays inserted into AT-rich dispersed elements. A comparative study by Southern analysis shows that these unusual dispersed elements containing the ZpS1 repeat are present with different organizations in all investigated Zamia species. In some species these elements are present with a low copy number, while in other species secondary amplification events, involving specific sequence clusters, appear to have generated characteristic dispersed elements in a high copy number. Among Zamia species, several groups share similar restriction patterns, as the Zamia loddigesii complex and the Caribbean species suggesting a general correlation between organization and genomic representation of the dispersed repeated sequence and the pattern of phyletic relationships in the genus. -

Ecological Sorting of Vascular Plant Classes During the Paleozoic Evolutionary Radiation
i1 Ecological Sorting of Vascular Plant Classes During the Paleozoic Evolutionary Radiation William A. DiMichele, William E. Stein, and Richard M. Bateman DiMichele, W.A., Stein, W.E., and Bateman, R.M. 2001. Ecological sorting of vascular plant classes during the Paleozoic evolutionary radiation. In: W.D. Allmon and D.J. Bottjer, eds. Evolutionary Paleoecology: The Ecological Context of Macroevolutionary Change. Columbia University Press, New York. pp. 285-335 THE DISTINCTIVE BODY PLANS of vascular plants (lycopsids, ferns, sphenopsids, seed plants), corresponding roughly to traditional Linnean classes, originated in a radiation that began in the late Middle Devonian and ended in the Early Carboniferous. This relatively brief radiation followed a long period in the Silurian and Early Devonian during wrhich morphological complexity accrued slowly and preceded evolutionary diversifications con- fined within major body-plan themes during the Carboniferous. During the Middle Devonian-Early Carboniferous morphological radiation, the major class-level clades also became differentiated ecologically: Lycopsids were cen- tered in wetlands, seed plants in terra firma environments, sphenopsids in aggradational habitats, and ferns in disturbed environments. The strong con- gruence of phylogenetic pattern, morphological differentiation, and clade- level ecological distributions characterizes plant ecological and evolutionary dynamics throughout much of the late Paleozoic. In this study, we explore the phylogenetic relationships and realized ecomorphospace of reconstructed whole plants (or composite whole plants), representing each of the major body-plan clades, and examine the degree of overlap of these patterns with each other and with patterns of environmental distribution. We conclude that 285 286 EVOLUTIONARY PALEOECOLOGY ecological incumbency was a major factor circumscribing and channeling the course of early diversification events: events that profoundly affected the structure and composition of modern plant communities. -

Estado De Conservación De Zamia Prasina W. Bull: Factores Históricos Que Incidieron En Su Diversidad Genética Y Demografia
Centro de Investigación Científica de Yucatán, A.C. Posgrado en Ciencias Biológicas ESTADO DE CONSERVACIÓN DE ZAMIA PRASINA W. BULL: FACTORES HISTÓRICOS QUE INCIDIERON EN SU DIVERSIDAD GENÉTICA Y DEMOGRAFIA. Tesis que presenta GRECIA MONTALVO FERNÁNDEZ En opción al título de DOCTOR EN CIENCIAS BIOLÓGICAS Opción recursos naturales Mérida, Yucatán, México 2020 DECLARACIÓN DE PROPIEDAD Declaro que la información contenida en la sección de Materiales y Métodos Experimentales, los Resultados y Discusión de este documento proviene de las actividades de experimentación realizadas durante el período que se me asignó para desarrollar mi trabajo de tesis, en las Unidades y Laboratorios del Centro de Investigación Científica de Yucatán, A.C., y que a razón de lo anterior y en contraprestación de los servicios educativos o de apoyo que me fueron brindados, dicha información, en términos de la Ley Federal del Derecho de Autor y la Ley de la Propiedad Industrial, le pertenece patrimonialmente a dicho Centro de Investigación. Por otra parte, en virtud de lo ya manifestado, reconozco que de igual manera los productos intelectuales o desarrollos tecnológicos que deriven o pudieran derivar de lo correspondiente a dicha información, le pertenecen patrimonialmente al Centro de Investigación Científica de Yucatán, A.C., y en el mismo tenor, reconozco que si derivaren de este trabajo productos intelectuales o desarrollos tecnológicos, en lo especial, estos se regirán en todo caso por lo dispuesto por la Ley Federal del Derecho de Autor y la Ley de la Propiedad Industrial, en el tenor de lo expuesto en la presente Declaración. ________________________________ GRECIA MONTALVO FERNÁNDEZ Este trabajo se llevó a cabo en la Unidad de Recursos Naturales del Centro de Investigación Científica de Yucatán, y forma parte del proyecto titulado Estado de Conservación de Zamia prasina en la Provincia Biótica Península de Yucatán bajo la dirección del Dr Jaime Martínez Castillo. -
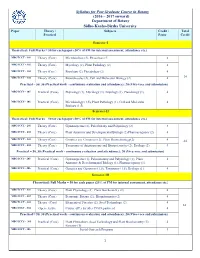
Syllabus for Post Graduate Course in Botany (2016 – 2017 Onward)
Syllabus for Post Graduate Course in Botany (2016 – 2017 onward) Department of Botany Sidho-Kanho-Birsha University Paper Theory / Subjects Credit / Total Practical Paper Credit Semester-I Theoretical: Full Marks = 50 for each paper (20% of FM for internal assessment, attendance etc.) MBOTCCT - 101 Theory (Core) Microbiology (2), Phycology (2) 4 MBOTCCT - 102 Theory (Core) Mycology (2), Plant Pathology (2) 4 MBOTCCT - 103 Theory (Core) Bryology (2), Pteridology (2) 4 MBOTCCT - 104 Theory (Core) Biomolecules (2), Cell and Molecular Biology (2) 4 24 Practical = 50, 30 (Practical work - continuous evaluation and attendance); 20 (Viva-voce and submission) MBOTCCS - 105 Practical (Core) Phycology (1), Mycology (1), Bryology (1), Pteridology (1). 4 MBOTCCS - 106 Practical (Core) Microbiology (1.5), Plant Pathology (1), Cell and Molecular 4 Biology (1.5). Semester-II Theoretical: Full Marks = 50 for each paper (20% of FM for internal assessment, attendance etc.) MBOTCCT - 201 Theory (Core) Gymnosperms (2), Paleobotany and Palynology (2) 4 MBOTCCT - 202 Theory (Core) Plant Anatomy and Developmental Biology (2) Pharmacognosy (2) 4 MBOTCCT - 203 Theory (Core) Genetics and Genomics (2), Plant Biotechnology(2) 4 24 MBOTCCT - 204 Theory (Core) Taxonomy of Angiosperms and Biosystematics (2), Ecology (2) 4 Practical = 50, 30 (Practical work - continuous evaluation and attendance); 20 (Viva-voce and submission) MBOTCCS - 205 Practical (Core) Gymnosperms (1), Palaeobotany and Palynology (1), Plant 4 Anatomy & Developmental Biology (1), Pharmacognosy (1). MBOTCCS - 206 Practical (Core) Genetics and Genomics (1.5), Taxonomy (1.5), Ecology (1). 4 Semester-III Theoretical: Full Marks = 50 for each paper (20% of FM for internal assessment, attendance etc.) MBOTCCT - 301 Theory (Core) Plant Physiology (2), Plant Biochemistry (2) 4 MBOTCCT - 302 Theory (Core) Economic Botany (2), Bioinformatics (2) 4 MBOTCCT - 303 Theory (Core) Elements of Forestry (2), Seed Technology (2). -

The Upper Carboniferous-Lower Permian Flora of Zöbing, Lower Austria
The upper Carboniferous-Lower Permian flora of Zöbing, Lower Austria Utrecht University Master Thesis Earth, Life and Climate By Koen Paalman 3470423 [email protected] 1 Contents page Abstract 3 1. Introduction 4 2. Geography, Geology and Lithology 6 3. Vegetation types 8 4. Different taxonomic groups 8 4.1 Calamites and other sphenopsids 8 4.2 Tree ferns 8 4.3 Cordaites 8 4.4 Pteridosperms 9 4.5 The medullosan pteridosperms 9 4.6 The peltaspermalean pteridosperms 9 5. Methods 10 6. Results 10 7. Correlation, comparison and interpretation 11 8. Discussion and conclusion 13 9. Acknowledgements 13 10. Appendix 14 10.1 Species list 14 10.2 Reference 16 11. Plates 19 2 Abstract During the Late Carboniferous and early Permian, a major floristic change took place in Euramerica. Gymnosperms replaced the previously dominant pteridophytes. This reflects a climatic change, i.e. from wetland-dominated to more arid conditions. Extensive studies on the vegetation during this time interval have recently been carried out in the Czech Republic. The Zöbing formation in Austria, is of the same age of these Czech formations, but has not yet been compared to them. Material from the Zöbing formation has been examined and compared to Czech floras. A clear transition can be seen in the flora of the Zöbing formation, from a Stephanian tree fern and Alethopteris zeilleri dominated flora, to a Asselian flora dominated by peltasperms and conifers. There are clear similarities between the floras of the Zöbing formation and the different Czechian formations, despite notable differences in species composition and abundance between Zöbing and the Czech formations. -

Curriculum Vitae
CURRICULUM VITAE ORCID ID: 0000-0003-0186-6546 Gar W. Rothwell Edwin and Ruth Kennedy Distinguished Professor Emeritus Department of Environmental and Plant Biology Porter Hall 401E T: 740 593 1129 Ohio University F: 740 593 1130 Athens, OH 45701 E: [email protected] also Courtesy Professor Department of Botany and PlantPathology Oregon State University T: 541 737- 5252 Corvallis, OR 97331 E: [email protected] Education Ph.D.,1973 University of Alberta (Botany) M.S., 1969 University of Illinois, Chicago (Biology) B.A., 1966 Central Washington University (Biology) Academic Awards and Honors 2018 International Organisation of Palaeobotany lifetime Honorary Membership 2014 Fellow of the Paleontological Society 2009 Distinguished Fellow of the Botanical Society of America 2004 Ohio University Distinguished Professor 2002 Michael A. Cichan Award, Botanical Society of America 1999-2004 Ohio University Presidential Research Scholar in Biomedical and Life Sciences 1993 Edgar T. Wherry Award, Botanical Society of America 1991-1992 Outstanding Graduate Faculty Award, Ohio University 1982-1983 Chairman, Paleobotanical Section, Botanical Society of America 1972-1973 University of Alberta Dissertation Fellow 1971 Paleobotanical (Isabel Cookson) Award, Botanical Society of America Positions Held 2011-present Courtesy Professor of Botany and Plant Pathology, Oregon State University 2008-2009 Visiting Senior Researcher, University of Alberta 2004-present Edwin and Ruth Kennedy Distinguished Professor of Environmental and Plant Biology, Ohio -

Diversity and Evolution of the Megaphyll in Euphyllophytes
G Model PALEVO-665; No. of Pages 16 ARTICLE IN PRESS C. R. Palevol xxx (2012) xxx–xxx Contents lists available at SciVerse ScienceDirect Comptes Rendus Palevol w ww.sciencedirect.com General palaeontology, systematics and evolution (Palaeobotany) Diversity and evolution of the megaphyll in Euphyllophytes: Phylogenetic hypotheses and the problem of foliar organ definition Diversité et évolution de la mégaphylle chez les Euphyllophytes : hypothèses phylogénétiques et le problème de la définition de l’organe foliaire ∗ Adèle Corvez , Véronique Barriel , Jean-Yves Dubuisson UMR 7207 CNRS-MNHN-UPMC, centre de recherches en paléobiodiversité et paléoenvironnements, 57, rue Cuvier, CP 48, 75005 Paris, France a r t i c l e i n f o a b s t r a c t Article history: Recent paleobotanical studies suggest that megaphylls evolved several times in land plant st Received 1 February 2012 evolution, implying that behind the single word “megaphyll” are hidden very differ- Accepted after revision 23 May 2012 ent notions and concepts. We therefore review current knowledge about diverse foliar Available online xxx organs and related characters observed in fossil and living plants, using one phylogenetic hypothesis to infer their origins and evolution. Four foliar organs and one lateral axis are Presented by Philippe Taquet described in detail and differ by the different combination of four main characters: lateral organ symmetry, abdaxity, planation and webbing. Phylogenetic analyses show that the Keywords: “true” megaphyll appeared at least twice in Euphyllophytes, and that the history of the Euphyllophytes Megaphyll four main characters is different in each case. The current definition of the megaphyll is questioned; we propose a clear and accurate terminology in order to remove ambiguities Bilateral symmetry Abdaxity of the current vocabulary. -

New Information on Bostonia Perplexa an Unusual Member of the Calamopityaceae from North America
Review o/ Palaeohotany and Pa(vnology, 72 ( 1992): 73 102 73 Elsevier Science Publishers B.V., Amsterdam New information on Bostonia perplexa an unusual member of the Calamopityaceae from North America William E. Stein, Jr. ~ and Charles B. Beck b ~Departmenl (?l Biolo~ical Sciences and Center Jbr Evolution and Paleoenvironment, State University (!/New York, Bin ghamton, N Y 13902-6000, USA bMuseum q/' Paleontology, University q[ Michigan, Ann Arbor, MI 48109, USA (Received September 25, 1991: revised and accepted January 15, 1992) ABSTRACT Stein, W.E. and Beck, C.B., 1992. New information on Bostonia pepTh'xa -an unusual member of the Calamopityaceae from North America. Rev. Palaeobot. Palynol., 72:73 102. A calamopityacean axis exhibiting multiple segments of primary xylem surrounded by secondary vascular tissue is analyzed here for its morphological and systematic significance. The plant is fundamentally protostelic with a deeply three-ribbed column of primary xylem. Each rib consists of a semi-discrete bundle of tracheids at the tip, intermittently connected to the stelar center by an extensive primary xylem parenchyma. The appearance of separate vascular segments at some levels is associated with departure of paired leaf traces. Between levels of trace departure, the three-ribbed protostele is reconstituted with primary xylem ribs following a helical course through the stem and supplying a regular Fibonacci phyllotaxis. Attached petiole bases are broadly of the Ka/vmma-type but exhibit a distinctly three-ribbed medial petiole bundle. The new specimen is assigned to Boslonia perph,xa requiring an expanded concept of the taxon. A restricted cladistic analysis of stelar architecture and nodal anatomy within the Calamopityaceae produces two phylogenetic hypotheses, One is preferred on morphological grounds but necessitates viewing at least some protostelic calamopityaceans as exhibiting a derived condition within the group.