Prediction of Molecular Crystal Structures
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-
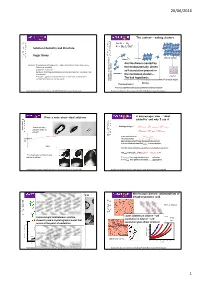
Roger Davey + Critical Nucleus Dimers Are the Dimers Created by Content: Studying and Defining Solute- Solute Interactions in the Liquid Phase
20/06/2016 The context – making clusters A + A A 2 K = [A ] / [A] 2 Solution Chemistry and Structure 2 Roger Davey + Critical nucleus dimers Are the dimers created by Content: Studying and defining solute- solute interactions in the liquid phase. Techniques available. thermodynamically driven Speciation in solution. self association present in Do these (thermodynamic features have) an impact on nucleation rate tetramers or outcome? monomers the nucleation clusters – Examples – glycine/saccharin/benzoic acid/tetrolic acid/mandelic crystal acid/paminobenzoic acid/co-crystals. The link hypothesis . Equilibria in supersaturated homogeneous solution Formation and Growth of critical nuclei Kinetics Thermodynamics Are these equilibria and quasi equilibria in anyway related? Nucleation Summer School June 20-24 th 2016 University of Strathclyde Nucleation Summer School June 20-24 th 2016 University of Strathclyde First a note about ideal solutions A macroscopic view - ‘ideal solubility’ and why I use it. Enthalpy changes: The usual way: ∆Hsolution = ∆H sublimation + ∆H solvation dissolve solid in ∆H = ∆H + ∆H solvent solution fusion mixing In an ideal solution ∆H mixing = 0. compositi This means that (Es-s + Esolv-solv )/2 = Es-solv on Real solutions don’t have this exact balance and in a non-ideal solution ∆H mixing is thus non zero. time And the ideal solubility is given by a very familiar equation: ln(xideal )={ΔHF(1/T m-1/T)+ ΔCp[T m/T – ln(T m/T)-1]}/R The ideal way: melt solid and mix with solvent. If x > xideal then negative deviation …….solvation If x < xideal then positive deviation………aggregation. Nucleation Summer School June 20-24 th 2016 University of Strathclyde Nucleation Summer School June 20-24 th 2016 University of Strathclyde Macroscopic picture: polymorphism of Macroscopic picture: saccharin dihydroxybenzoic acid. -
Chemical Specificity in Short-Chain Fatty Acids and Their Analogues in Increasing Osmotic Fragility in Rat Erythrocytes in Vitro
Chemical specificity in short-chain fatty acids and their analogues in increasing osmotic fragility in rat erythrocytes in Title vitro. Author(s) Mineo, Hitoshi; Hara, Hiroshi Biochimica et Biophysica Acta (BBA) - Biomembranes, 1768(6), 1448-1453 Citation https://doi.org/10.1016/j.bbamem.2007.02.008 Issue Date 2007-06 Doc URL http://hdl.handle.net/2115/28208 Type article (author version) File Information BBA1768-6.pdf Instructions for use Hokkaido University Collection of Scholarly and Academic Papers : HUSCAP 1 1 Chemical specificity in short-chain fatty acids and their analogues in increasing osmotic 2 fragility in rat erythrocytes in vitro 3 4 Hitoshi Mineo, Hiroshi Hara* 5 6 Division of Applied Bioscience, Graduate School of Agriculture, Hokkaido University, 7 Hokkaido 060-8589, Japan 8 9 10 *Corresponding author. 11 Hiroshi HARA Ph.D. 12 Division of Applied Bioscience, 13 Graduate School of Agriculture, 14 Hokkaido University, 15 Kita-9, Nishi-9, Sapporo, 16 Hokkaido 060-8589, 17 Tel.: +81-11-706-3352; 18 fax: +81-11-706-2504. 19 E-mail address: [email protected] 20 21 22 23 24 25 2 1 Abstract 2 3 We examined the role of the chemical specificity of short-chain fatty acids 4 (SCFAs) and their derivatives in increasing osmotic fragility (OF) in rat red blood cells 5 (RBCs). Except for formic acid, normal SCFAs with 2 to 8 carbons increased the OF in 6 rat RBCs with increasing number of hydrocarbons in a dose-dependent manner. 7 Replacement of the carboxylic group with sulfonic group inhibited, but did not abolish, 8 the SCFA-mediated increase in OF. -

United States Patent (19) 11 4,027,037 Siegle Et Al
United States Patent (19) 11 4,027,037 Siegle et al. (45) May 31, 1977 (54) N-SUBSTITUTED B-AMINOCROTONIC 57) ABSTRACT ACID ESTERS N-substituted (3-aminocrotonic acid esters of the for mula (75) Inventors: Peter Siegle, Cologne; Klaus Sasse, Schildgen; Peter Rössler, Cologne, all of Germany CH. COOR: (I) RC=CCN H (73) Assignee: Bayer Aktiengesellschaft, R11 Leverkusen, Germany in which (22 Filed: Mar. 12, 1975 R is hydrogen, or alkyl or alkenyl with up to 4 car bon atoms, (21) Appl. No.: 557,698 R’ is straight-chain or branched alkyl or alkenyl with up to 9 carbon atoms, optionally interrupted by O 30 Foreign Application Priority Data or S, and optionally substituted by halogen, phe Mar. 26, 1974 Germany .......................... 2414456 noxy, dimethylamino, cyclic alkyl or alkenyl with up to 8 carbon atoms which are optionally substi (52) U.S. C. ......................... 424/314; 260/239 BE; tuted by halogen, dimethylamino, or alkyl, alkoxy 260/239 BF; 260/247.2 B; 260/268 R; or alkylthio with up to 4 carbon atoms, phenyl 260/293.88; 260/326.43; 260/340.5; which is optionally substituted by halogen, dimeth 260/347.4; 260/468 G; 260/468 J; 260/470; ylamino, phenoxy, or alkyl, alkenyl, acyl, alkoxy, 260/471 A; 260/479 S; 260/481 R; 260/482 alkenoxy or alkynoxy containing up to 4 carbon R; 424/244; 424/2 48.52; 424/250; 424/267; atoms, or a 5- or 6-membered heterocyclic ring 424/274; 424/282; 424/285; 424/299; containing at least one O, S or N hetero-atom and 424/309; 424/248.55 optionally substituted by halogen, alkyl or alkoxy containing up to 4 carbon atoms or R is further (51 int. -

(12) United States Patent (10) Patent No.: US 8,623.422 B2 Hansen Et Al
USOO8623422B2 (12) United States Patent (10) Patent No.: US 8,623.422 B2 Hansen et al. (45) Date of Patent: *Jan. 7, 2014 (54) COMBINATION TREATMENT WITH 5,668,161 A 9/1997 Talley et al. STRONTUM FOR THE PROPHYLAXIS 5,681,842 A 10, 1997 Dellaria et al. AND/OR TREATMENT OF CARTILAGE 5,686.460 A 11/1997 Nicolai et al. AND/OR BONE CONDITIONS 5,686,470 A 11/1997 Weier et al. 5,696,431 A 12/1997 Giannopoulos et al. 5,707,980 A 1/1998 Knutson (75) Inventors: Christian Hansen, Vedback (DK); 5,719,163 A 2f1998 Norman et al. Henrik Nilsson, Copenhagen (DK); 5,750,558 A 5/1998 Brooks et al. Stephan Christgau, Gentofte (DK) 5,753,688 A 5/1998 Talley et al. 5,756,530 A 5/1998 Lee et al. (73) Assignee: Osteologix A/S, Copenhagen (DK) 5,756,531 A 5/1998 Brooks et al. 5,760,068 A 6/1998 Talley et al. (*) Notice: Subject to any disclaimer, the term of this 5,776,967 A 7/1998 Kreft et al. patent is extended or adjusted under 35 5,776,984. A 7/1998 Dellaria et al. U.S.C. 154(b) by 558 days. 5,783,597 A 7/1998 Beers et al. This patent is Subject to a terminal dis 5,807,873 A 9, 1998 Nicolai et al. claimer. 5,824,699 A 10, 1998 Kreft et al. 5,830,911 A 11/1998 Failli et al. 5,840,924 A 11/1998 Desmond et al. -

Patent Office Paiented May 2, 1963
United States Patent Office Paiented May 2, 1963 2 In addition, these prior metalation processes are gen 3,999,859 erally deemed unsuitable for commercialization due to TANSFETALATON PRESCESS Water E. Foster, Batca Rouge, La., assiggor to Eainy the difficulties and high costs connected with their use. Corporation, New York, N.Y., a corporation of Further, the prior processes require many separate proc Virginia eSS Steps as well as long periods for appreciable reaction No Draving. Fied Feb. 24, 1959, Ser. No. 794,817 which at best converts only half of the reactants, e.g., A2 Cains. (C. 262-665) toluene and sodium, to the desired product and com pletely degrades the chlorine values to sodium chloride. This invention relates to a method of preparing organic The above objectionable features of the prior art mate compounds and more particularly to the preparation of O rialiy increase the overall costs of the desired end prod aliphatic polyolefin hydrocarbons and organo alkali metal uct, and nake its manufacture by these processes com compounds. mercially unattractive. Aliphatic polyolefin hydrocarbons are extremely inter it is accordingly an object of the present invention to esting chemical compounds which are readily converted provide a process for the manufacture of aliphatic poly into valuable industrial materials by processes well known 5 olefin hydrocarbons. Another object is to provide a proc to those skilled in the art. Of particular interest are eSS of the above type which is suitable for manufacturing those long chain aliphatic polyolefin compounds where a very wide variety of aliphatic polyolefin hydrocarbons, in the unsaturation is located near the terminal carbons including many which were heretofore unknown or not of the molecular chain. -

Organic Reactions VOLUME IV
Organic Reactions VOLUME IV EDITORIAL BOARD ROGER ADAMS, Editor-in-Chief WERNER E. BACHMANN LOUIS F. FIESER A. H. BLATT JOHN R. JOHNSON HAROLD R. SNYDER ASSOCIATE EDITORS JAMES CASON MILTON C. KLOETZEL WILLIAM S. EMERSON S. M. MCELVAIN H. L. HOLMES ERICH MOSETTIG WALTER S. IDE DAVID TODD NEW YORK JOHN WILEY & SONS, INC. LONDON: CHAPMAN & HALL, LIMITED COPYRIGHT, 1948 BY ROGER ADAMS All Rights Reserved This book or any part thereof must not be reproduced in any form without the written permission of the ptihlisher. PRINTED IN THE UNITED STATES OF AMERICA PREFACE TO THE SERIES In the course of nearly every program of research in organic chemistry the investigator finds it necessary to use several of the better-known synthetic reactions. To discover the optimum conditions for the appli cation of even the most familiar one to a compound not previously subjected to the reaction often requires an extensive search of the liter ature; even then a series of experiments may be necessary. When the results of the investigation are published, the synthesis, which may have required months of work, is usually described without comment. The background of knowledge and experience gained in the literature search and experimentation is thus lost to those who subsequently have occasion to apply the general method. The student of preparative organic chemistry faces similar difficulties. The textbooks and labora tory manuals furnish numerous examples of the application of various syntheses, but only rarely do they convey an accurate conception of the scope and usefulness of the processes. For many years American organic chemists have discussed these problems. -

Plain Paper:909Aiche.P
Materials, Interfaces, and Electrochemical Phenomena A New Technique for Predicting the Shape of Solution-Grown Organic Crystals Daniel Winn and Michael F. Doherty Dept. of Chemical Engineering, University of Massachusetts, Amherst, MA 01003 A method for predicting the shape of solution-grown organic crystals is presented. The shapes depend on properties of their internal crystal structure, as well as on the processing en®ironment. The model de®eloped here can account for the effects of sol- ®ent and may be extended to account for the influence of certain types of additi®es. This method may be used as a first approach for including crystal shape in the o®erall design and optimization of organic solids] processes. The technique has been used successfully to predict the shape of adipic acid grown from water, biphenyl from toluene, and ibuprofen from polar and nonpolar sol®ents. Introduction The shape of crystalline organic solids is an important fac- actions between organic molecules in the solid state has led tor in the design and operation of solid]liquid separation sys- to a burgeoning of ideas of how these forces influence mor- tems. Crystal morphology affects the efficiency of down- phology. Saska and MyersonŽ. 1983 and Berkowitch-Yellen stream processesŽ. such as filtering, washing, and drying , and Ž.1985 were among the first to calculate solid-state energies influences material properties such as bulk density and me- of organic materials for the purpose of morphological model- chanical strength which play a major role in storage and han- ing. An especially important contribution has been the work dling. -

I. Intramolecular Diels-Alder Cycloadditions of Citraconic and Mesaconic Esters II. Studies on the Synthesis of the Northwest An
AN ABSTRACT OF THE THESIS OF Bernard Gale Sheldon for the degree of Doctor of Philosophy in Chemistry presented on October 17, 1980 Title: I. INTRAMOLECULAR DIELS-ALDER CYCLOADDITIONS OF CITRACONIC AND MESACONIC ESTERS II. STUDIES ON THE SYNTHESIS OF THE NORTHWEST AND SOUTHEAST SEGMENTS OF BOROMYCIN Abstract approved: Redacted for privacy Dr. James D. White The thermal cycloaddition of trans,trans-2,4-hexadienyl 3-carboxy- crotonate 95 was found to produce cis-3a-hydroxymethy1-18,6a-dimethyl- cyclohex-4-ene-la,2a-dicarboxylic acid y-lactone (97), the structure of which was confirmed by reduction of the methyl ester 98, with diisobutyl- aluminum hydride to the acetal 99, exo-l-endo-10-dimethy1-3,5-dioxatricyclo 4,11 [5.3.1.0 ] undec-8-ene. An X-ray crystallographic study further con- firmed structure 97. Trans,trans-2,4-hexadienyl trans-3-methoxycarbonyl- 3-methylacrylate 90 was found to undergo cycloaddition to yield a 4.3:1 mixture of trans-3a-hydroxymethy1-18,6a-dimethyl-la-methoxycarbonyl- cyclohex-4-ene-2f3-carboxylic acid y-lactone (117) and cis-3a-hydroxymethyl- la,6a-dimethy1-18-methoxycarbonylcyclohex-4-ene-2a-carboxylic acid 'y- lactone (113). Epimerization of 117 with sodium hydride produced 98. The citraconic ester 110, trans,trans-2,4-hexadienyl 3-methoxycarbonyl- crotonate, was thermally cyclized to trans-3a-hydroxymethyl-la,6a- dimethyl-l-methoxycarbonylcyclohex-4-ene-2a-carboxylic acid y-lactone (112), which could be epimerized with sodium methoxide to cis-3a- hydroxymethyl-la,6a-dimethy1-18,-methoxycarbonyl-cyclohex-4-ene-2a- carboxylic acid y-lactone (113). -

Modeling Crystal Shapes of Organic Materials Grown from Solution
SEPARATIONS JOURNAL REVIEW Modeling Crystal Shapes of Organic Materials Grown from Solution Daniel Winn and Michael F. Doherty Dept. of Chemical Engineering, University of Massachusetts, Amherst, MA 01003 The shape of a crystalline organic solid has a major impact on its downstream pro- cessing and on its end-product quality, issues that are becoming increasingly important in the specialty and fine chemical, as well as the pharmaceutical and life science, indus- tries. Though it is widely known that impro®ed crystal shapes can be achie®ed by ®ary- ing the conditions of crystallization() such as sol®ent type and impurity le®els , there is far less understanding of how to effect such a change. Until recently, most methods for predicting crystal shapes were based exclusi®ely on the internal crystal structure, and hence could not account for sol®ent or impurity effects. New approaches, howe®er, offer the possibility of accurately predicting the effects of sol®ents. Models for predicting crys- tal shape are re®iewed, as well as their utility for process and product design. Introduction In the chemical process industries, numerous organic ma- shape must be controlled for both medical and regulatory terials are purified by solid-liquid separationŽ such as adipic purposesŽ. Romero et al., 1991 . Crystal morphology also af- acid, ibuprofen, and bisphenol A. Many of them, in particu- fects the ease with which the drug is compressed into tablets lar specialty chemicals such as pharmaceuticals, are crystal- Ž.Gordon and Amin, 1984 . Both are key factors in the pro- lized from solution. As with other separation techniques, the cess efficiency and product quality of pharmaceuticalsŽ York, product purity is the primary measure of product quality. -
GFS Organics Modern Acetylene Chemistry for Research and Applied Technology ORDERING INFORMATION
GFS ORGANIC® CHEMICALSGFS Organics Modern Acetylene Chemistry for Research and Applied Technology ORDERING INFORMATION Customer Service Representatives and Technical Support are available Monday through Friday from 8:00 a.m. to 5:30 p.m. Eastern Time. Orders may be placed by mail, phone, fax, e-mail or on our web site. Every order receives an order confirmation sent via fax or e-mail that includes items ordered, pricing, shipping method, and estimated shipping date. By Mail: GFS CHEMICALS, INC. 800 Kaderly Drive Columbus, OH 43228 By Phone: 800-858-9682 (U.S. and Canada) (740) 881-5501 (International) By Fax: (740) 881-5989 By E-mail: [email protected] For Applied Technologies, R&D, or Pilot Scale email: [email protected] On the Web: www.gfschemicals.com www.gfsinternational.com Opening an account GFS Customer Service Representatives will be glad to help you open an account. For details call: 1-800-858-9682. (740) 881-5501 (International) For full terms and conditions visit http://www.gfschemicals.com/statics/ documents/aboutus/termsandconditions.html Notice: GFS products are not certified or intended for direct use in food, drug, or cosmetic applications; they are warranted to meet our specifica- tions as given in GFS Lot Analysis or other Company literature. Determi- nation of material suitability and/or merchantability for purposes beyond these specifications is the responsibility of the end user. We Honor WHY SOURCE FROM GFS? GFS is recognized as a world Customers benefit from our A Sampling of Our Core leader in the production of knowledge and expertise Organic Chemistries alkynes and downstream in the formulation of spe- derivative technology. -

High Oleic Polyol Esters, Compositions and Lubricants Functional Fluids and Greases Containing the Same
Europaisches Patentamt European Patent Office Office europeen des brevets (11) EP 0 712 834 A1 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) |nt CI C07C 69/52, C10M 105/38, 22.05.1996 Bulletin 1996/21 C07C Q7/Q2, C10M 129/10, C10M 133/12, C10M 133/40 (21)to 1 \ Applicationa , ♦■ number:k 95308145.2oc,nfi1«, ' (22) Date of filing: 14.11.1995 (84) Designated Contracting States: • Lai, Kasturi BE DE ES FR GB IT NL SE Euclid, Ohio 44123 (US) (30) Priority: 15.11.1994 US 339821 (74) Representative: Crisp, David Norman et al D. YOUNG & CO. (71) Applicant: THE LUBRIZOL CORPORATION 21 New Fetter Lane Wickliffe, Ohio 44092-2298 (US) London EC4A 1 DA (GB) (72) Inventors: • Lawate, Saurabh Shripad Concord, Ohio 44060 (US) (54) High oleic polyol esters, compositions and lubricants functional fluids and greases containing the same (57) A polyol ester is described which is derived The invention also relates to compositions comprising from: the polyol esters and at least one antioxidant, and lubri- cating oil compositions comprising an oil of lubricating (A) an aliphatic or alicyclic polyol: and viscosity and the polyol esters of the invention. Lubricat- ing oil compositions comprising polyol esters of the in- (B) an aliphatic monocarboxylic mixture derived vention, at least one antioxidant, and an oil of lubricating from a natural vegetable oil, said acid mixture com- viscosity also are described and are particularly useful. prising at least about 72% by weight of oleic acid. ^- CO 00 CM o a. LU Printed by Jouve, 75001 PARIS (FR) EP 0 712 834 A1 Description The invention relates to polyol esters, and more particularly, to polyol esters derived from polyols and aliphatic monocarboxylic acid mixtures derived from natural vegetable oils wherein the acid mixtures comprise at least about 5 72% by weight of oleic acid. -

Chemical Specificity in Short-Chain Fatty Acids and Their Analogues in Increasing Osmotic Fragility in Rat Erythrocytes in Vitro ⁎ Hitoshi Mineo, Hiroshi Hara
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Biochimica et Biophysica Acta 1768 (2007) 1448–1453 www.elsevier.com/locate/bbamem Chemical specificity in short-chain fatty acids and their analogues in increasing osmotic fragility in rat erythrocytes in vitro ⁎ Hitoshi Mineo, Hiroshi Hara Division of Applied Bioscience, Graduate School of Agriculture, Hokkaido University, Kita-9, Nishi-9, Sapporo, Hokkaido 060-8589, Japan Received 30 October 2006; received in revised form 7 February 2007; accepted 7 February 2007 Available online 17 February 2007 Abstract We examined the role of the chemical specificity of short-chain fatty acids (SCFAs) and their derivatives in increasing osmotic fragility (OF) in rat red blood cells (RBCs). Except for formic acid, normal SCFAs with 2 to 8 carbons increased the OF in rat RBCs with increasing number of hydrocarbons in a dose-dependent manner. Replacement of the carboxylic group with sulfonic group inhibited, but did not abolish, the SCFA- mediated increase in OF. Introduction of another carboxylic group (dicarboxylic acids) completely abolished the SCFA-mediated increase in OF. Transformation of the hydrocarbon chains in SCFAs from straight to branched or cyclic chains affected the degree of the OF-increasing effect. Introduction of double- or triple-carbon bonds to the hydrocarbon chain in parent SCFAs did not affect the increase in OF. Both hydrophilic (carboxylic group) and hydrophobic elements (hydrocarbons) at opposite sides of a molecule were required to affect the RBC membrane, and the size and form of hydrophobic element were important factors in determining the SCFA-mediated increase in OF.