(12) Patent Application Publication (10) Pub. No.: US 2009/0297536A1 Chin Et Al
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Efficacy and Mechanistic Evaluation of Tic10, a Novel Antitumor Agent
University of Pennsylvania ScholarlyCommons Publicly Accessible Penn Dissertations 2012 Efficacy and Mechanisticv E aluation of Tic10, A Novel Antitumor Agent Joshua Edward Allen University of Pennsylvania, [email protected] Follow this and additional works at: https://repository.upenn.edu/edissertations Part of the Oncology Commons Recommended Citation Allen, Joshua Edward, "Efficacy and Mechanisticv E aluation of Tic10, A Novel Antitumor Agent" (2012). Publicly Accessible Penn Dissertations. 488. https://repository.upenn.edu/edissertations/488 This paper is posted at ScholarlyCommons. https://repository.upenn.edu/edissertations/488 For more information, please contact [email protected]. Efficacy and Mechanisticv E aluation of Tic10, A Novel Antitumor Agent Abstract TNF-related apoptosis-inducing ligand (TRAIL; Apo2L) is an endogenous protein that selectively induces apoptosis in cancer cells and is a critical effector in the immune surveillance of cancer. Recombinant TRAIL and TRAIL-agonist antibodies are in clinical trials for the treatment of solid malignancies due to the cancer-specific cytotoxicity of TRAIL. Recombinant TRAIL has a short serum half-life and both recombinant TRAIL and TRAIL receptor agonist antibodies have a limited capacity to perfuse to tissue compartments such as the brain, limiting their efficacy in certain malignancies. To overcome such limitations, we searched for small molecules capable of inducing the TRAIL gene using a high throughput luciferase reporter gene assay. We selected TRAIL-inducing compound 10 (TIC10) for further study based on its induction of TRAIL at the cell surface and its promising therapeutic index. TIC10 is a potent, stable, and orally active antitumor agent that crosses the blood-brain barrier and transcriptionally induces TRAIL and TRAIL-mediated cell death in a p53-independent manner. -

Tpd52l2 (NM 025482) Mouse Tagged ORF Clone – MR202509
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for MR202509 Tpd52l2 (NM_025482) Mouse Tagged ORF Clone Product data: Product Type: Expression Plasmids Product Name: Tpd52l2 (NM_025482) Mouse Tagged ORF Clone Tag: Myc-DDK Symbol: Tpd52l2 Synonyms: 2810411G23Rik; AU016537; C86069; D54 Vector: pCMV6-Entry (PS100001) E. coli Selection: Kanamycin (25 ug/mL) Cell Selection: Neomycin ORF Nucleotide >MR202509 ORF sequence Sequence: Red=Cloning site Blue=ORF Green=Tags(s) TTTTGTAATACGACTCACTATAGGGCGGCCGGGAATTCGTCGACTGGATCCGGTACCGAGGAGATCTGCC GCCGCGATCGCC ATGGACTCTGCTAGCCAAGATATCAACCTGAATTCTCCTAACAAAGGTGTGCTGTCTGACTTTATGACTG ACGTCCCTGTTGACCCAGGTGTGGTCCACCGGACTCCAGTTGTAGAAGGCCTGACAGAGGGGGAGGAAGA AGAGCTTCGGGCTGAGCTTGCTAAGGTGGAAGAAGAAATTGTCACTCTGCGCCAGGTGCTGGCAGCCAAA GAGAGGCACTGTGGAGAGCTGAAAAGGAGGCTGGGCCTCTCCACATTAGGGGAGCTGAAGCAGAACCTGT CTAGGAGCTGGCATGATGTGCAGGTCTCTACTGCCTATGTGAAAACGTCTGAGAAACTTGGAGAGTGGAA TGAGAAAGTGACGCAGTCTGACCTCTACAAGAAGACTCAAGAAACTCTTTCACAGGCTGGACAGAAAACA TCAGCTGCCCTGTCCACCATGGGCTCTGCCATCAGCAGGAAGCTTGGAGACATGAGCAGCTACTCCATCC GCCACTCGATAAGTATGCCTGTCATGAGGAACTCAGCCACCTTCAAGTCATTTGAAGACCGAGTGGGGAC CATAAAGTCTAAGGTTGTGGGTGGCAGAGAGAATGGCAGCGATAACCTCCCTCCCTCTCCTGGAAGTGGT GACCAGACATTGCCGGATCATGCGCCTTTC ACGCGTACGCGGCCGCTCGAGCAGAAACTCATCTCAGAAGAGGATCTGGCAGCAAATGATATCCTGGATT ACAAGGATGACGACGATAAGGTTTAA This product is to be used for laboratory only. Not for diagnostic or therapeutic use. -

Identification of Tumor-Associated Cassette Exons in Human Cancer
Valletti et al. Molecular Cancer 2010, 9:230 http://www.molecular-cancer.com/content/9/1/230 RESEARCH Open Access Identification of tumor-associated cassette exons in human cancer through EST-based computational prediction and experimental validation Alessio Valletti1†, Anna Anselmo2†, Marina Mangiulli1, Ilenia Boria1, Flavio Mignone3, Giuseppe Merla4, Vincenzo D’Angelo5, Apollonia Tullo6, Elisabetta Sbisà6, Anna Maria D’Erchia1, Graziano Pesole1,7* Abstract Background: Many evidences report that alternative splicing, the mechanism which produces mRNAs and proteins with different structures and functions from the same gene, is altered in cancer cells. Thus, the identification and characterization of cancer-specific splice variants may give large impulse to the discovery of novel diagnostic and prognostic tumour biomarkers, as well as of new targets for more selective and effective therapies. Results: We present here a genome-wide analysis of the alternative splicing pattern of human genes through a computational analysis of normal and cancer-specific ESTs from seventeen anatomical groups, using data available in AspicDB, a database resource for the analysis of alternative splicing in human. By using a statistical methodology, normal and cancer-specific genes, splice sites and cassette exons were predicted in silico. The condition association of some of the novel normal/tumoral cassette exons was experimentally verified by RT-qPCR assays in the same anatomical system where they were predicted. Remarkably, the presence in vivo of the predicted alternative transcripts, specific for the nervous system, was confirmed in patients affected by glioblastoma. Conclusion: This study presents a novel computational methodology for the identification of tumor-associated transcript variants to be used as cancer molecular biomarkers, provides its experimental validation, and reports specific biomarkers for glioblastoma. -
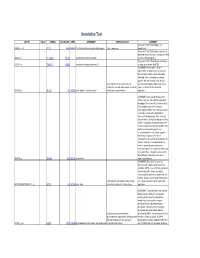
Supplementary Table 5. Functional Annotation of the Largest Gene Cluster(221 Element)
Annotation Tool AFFYID VALUE SYMBOL LOCUSLINK OMIM GENENAME GENEONTOLOGY SUMMARY [Proteome FUNCTION:] Expressed 203054_s_at TCTA 6988 600690 T-cell leukemia translocation altered gene tumor suppressor ubiquitously [Proteome FUNCTION:] May be involved in protein-protein interactions; contains five WD 44563_at FLJ10385 55135 hypothetical protein FLJ10385 domains (WD-40 repeats) [Proteome FUNCTION:] Weakly similarity to 212261_at TNRC15 26058 trinucleotide repeat containing 15 a region of rat nestin (Rn.9701) [SUMMARY:] Actin alpha 1 which is expressed in skeletal muscle is one of six different actin isoforms which have been identified. Actins are highly conserved proteins that are involved in cell motility, actin filament; motor activity; muscle structure and integrity. Alpha actins are a contraction; muscle development; structural major constituent of the contractile 203872_at ACTA1 58 102610 actin, alpha 1, skeletal muscle constituent of cytoskeleton apparatus. [SUMMARY:] Annexin VIII belong to the family of Ca (2+) dependent phospholipid binding proteins (annexins), and has a high 56% identity to annexin V (vascular anticoagulant-alpha). It was initially isolated as 2.2 kb vascular anticoagulant-beta transcript from human placenta, a Ca (2+) dependent phospholipid binding protein that inhibits coagulation and phospholipase A2 activity. However, the fact that annexin VIII is neither an extracellular protein nor associated with the cell surface suggests that it may not play a role in blood coagulation in vivo and its physiological role remains unknown. It is expressed at low levels in human placenta and shows restricted expression in lung endothelia, skin, liver, and kidney. The gene is also found to be selectively overexpressed in acute 203074_at ANXA8 244 602396 annexin A8 myelocytic leukemia. -

Supplementary Materials
Supplementary materials Supplementary Table S1: MGNC compound library Ingredien Molecule Caco- Mol ID MW AlogP OB (%) BBB DL FASA- HL t Name Name 2 shengdi MOL012254 campesterol 400.8 7.63 37.58 1.34 0.98 0.7 0.21 20.2 shengdi MOL000519 coniferin 314.4 3.16 31.11 0.42 -0.2 0.3 0.27 74.6 beta- shengdi MOL000359 414.8 8.08 36.91 1.32 0.99 0.8 0.23 20.2 sitosterol pachymic shengdi MOL000289 528.9 6.54 33.63 0.1 -0.6 0.8 0 9.27 acid Poricoic acid shengdi MOL000291 484.7 5.64 30.52 -0.08 -0.9 0.8 0 8.67 B Chrysanthem shengdi MOL004492 585 8.24 38.72 0.51 -1 0.6 0.3 17.5 axanthin 20- shengdi MOL011455 Hexadecano 418.6 1.91 32.7 -0.24 -0.4 0.7 0.29 104 ylingenol huanglian MOL001454 berberine 336.4 3.45 36.86 1.24 0.57 0.8 0.19 6.57 huanglian MOL013352 Obacunone 454.6 2.68 43.29 0.01 -0.4 0.8 0.31 -13 huanglian MOL002894 berberrubine 322.4 3.2 35.74 1.07 0.17 0.7 0.24 6.46 huanglian MOL002897 epiberberine 336.4 3.45 43.09 1.17 0.4 0.8 0.19 6.1 huanglian MOL002903 (R)-Canadine 339.4 3.4 55.37 1.04 0.57 0.8 0.2 6.41 huanglian MOL002904 Berlambine 351.4 2.49 36.68 0.97 0.17 0.8 0.28 7.33 Corchorosid huanglian MOL002907 404.6 1.34 105 -0.91 -1.3 0.8 0.29 6.68 e A_qt Magnogrand huanglian MOL000622 266.4 1.18 63.71 0.02 -0.2 0.2 0.3 3.17 iolide huanglian MOL000762 Palmidin A 510.5 4.52 35.36 -0.38 -1.5 0.7 0.39 33.2 huanglian MOL000785 palmatine 352.4 3.65 64.6 1.33 0.37 0.7 0.13 2.25 huanglian MOL000098 quercetin 302.3 1.5 46.43 0.05 -0.8 0.3 0.38 14.4 huanglian MOL001458 coptisine 320.3 3.25 30.67 1.21 0.32 0.9 0.26 9.33 huanglian MOL002668 Worenine -

(P -Value<0.05, Fold Change≥1.4), 4 Vs. 0 Gy Irradiation
Table S1: Significant differentially expressed genes (P -Value<0.05, Fold Change≥1.4), 4 vs. 0 Gy irradiation Genbank Fold Change P -Value Gene Symbol Description Accession Q9F8M7_CARHY (Q9F8M7) DTDP-glucose 4,6-dehydratase (Fragment), partial (9%) 6.70 0.017399678 THC2699065 [THC2719287] 5.53 0.003379195 BC013657 BC013657 Homo sapiens cDNA clone IMAGE:4152983, partial cds. [BC013657] 5.10 0.024641735 THC2750781 Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). 4.07 0.04353262 DNAH5 [Source:Uniprot/SWISSPROT;Acc:Q8TE73] [ENST00000382416] 3.81 0.002855909 NM_145263 SPATA18 Homo sapiens spermatogenesis associated 18 homolog (rat) (SPATA18), mRNA [NM_145263] AA418814 zw01a02.s1 Soares_NhHMPu_S1 Homo sapiens cDNA clone IMAGE:767978 3', 3.69 0.03203913 AA418814 AA418814 mRNA sequence [AA418814] AL356953 leucine-rich repeat-containing G protein-coupled receptor 6 {Homo sapiens} (exp=0; 3.63 0.0277936 THC2705989 wgp=1; cg=0), partial (4%) [THC2752981] AA484677 ne64a07.s1 NCI_CGAP_Alv1 Homo sapiens cDNA clone IMAGE:909012, mRNA 3.63 0.027098073 AA484677 AA484677 sequence [AA484677] oe06h09.s1 NCI_CGAP_Ov2 Homo sapiens cDNA clone IMAGE:1385153, mRNA sequence 3.48 0.04468495 AA837799 AA837799 [AA837799] Homo sapiens hypothetical protein LOC340109, mRNA (cDNA clone IMAGE:5578073), partial 3.27 0.031178378 BC039509 LOC643401 cds. [BC039509] Homo sapiens Fas (TNF receptor superfamily, member 6) (FAS), transcript variant 1, mRNA 3.24 0.022156298 NM_000043 FAS [NM_000043] 3.20 0.021043295 A_32_P125056 BF803942 CM2-CI0135-021100-477-g08 CI0135 Homo sapiens cDNA, mRNA sequence 3.04 0.043389246 BF803942 BF803942 [BF803942] 3.03 0.002430239 NM_015920 RPS27L Homo sapiens ribosomal protein S27-like (RPS27L), mRNA [NM_015920] Homo sapiens tumor necrosis factor receptor superfamily, member 10c, decoy without an 2.98 0.021202829 NM_003841 TNFRSF10C intracellular domain (TNFRSF10C), mRNA [NM_003841] 2.97 0.03243901 AB002384 C6orf32 Homo sapiens mRNA for KIAA0386 gene, partial cds. -

The DNA Sequence and Comparative Analysis of Human Chromosome 20
articles The DNA sequence and comparative analysis of human chromosome 20 P. Deloukas, L. H. Matthews, J. Ashurst, J. Burton, J. G. R. Gilbert, M. Jones, G. Stavrides, J. P. Almeida, A. K. Babbage, C. L. Bagguley, J. Bailey, K. F. Barlow, K. N. Bates, L. M. Beard, D. M. Beare, O. P. Beasley, C. P. Bird, S. E. Blakey, A. M. Bridgeman, A. J. Brown, D. Buck, W. Burrill, A. P. Butler, C. Carder, N. P. Carter, J. C. Chapman, M. Clamp, G. Clark, L. N. Clark, S. Y. Clark, C. M. Clee, S. Clegg, V. E. Cobley, R. E. Collier, R. Connor, N. R. Corby, A. Coulson, G. J. Coville, R. Deadman, P. Dhami, M. Dunn, A. G. Ellington, J. A. Frankland, A. Fraser, L. French, P. Garner, D. V. Grafham, C. Grif®ths, M. N. D. Grif®ths, R. Gwilliam, R. E. Hall, S. Hammond, J. L. Harley, P. D. Heath, S. Ho, J. L. Holden, P. J. Howden, E. Huckle, A. R. Hunt, S. E. Hunt, K. Jekosch, C. M. Johnson, D. Johnson, M. P. Kay, A. M. Kimberley, A. King, A. Knights, G. K. Laird, S. Lawlor, M. H. Lehvaslaiho, M. Leversha, C. Lloyd, D. M. Lloyd, J. D. Lovell, V. L. Marsh, S. L. Martin, L. J. McConnachie, K. McLay, A. A. McMurray, S. Milne, D. Mistry, M. J. F. Moore, J. C. Mullikin, T. Nickerson, K. Oliver, A. Parker, R. Patel, T. A. V. Pearce, A. I. Peck, B. J. C. T. Phillimore, S. R. Prathalingam, R. W. Plumb, H. Ramsay, C. M. -

Perspectives
FOCUS ON MECHANOTRANSDUCTION PERSPECTIVES network that can promote coordinated OPINION changes in cell, cytoskeletal and nuclear struc- ture in response to mechanical distortion14 Mechanotransduction at a (FIG. 1a). (Herein, the term hard-wired refers to cytoskeletal structures that are stable enough distance: mechanically coupling the as interconnected units to resist mechanical stresses and thereby maintain shape stabil- ity, even though they undergo continuous extracellular matrix with the nucleus dynamic remodelling at the molecular level.) This model takes into account the observa- Ning Wang, Jessica D. Tytell and Donald E. Ingber tion that individual cytoskeletal filaments can bear significant tensile and compressive loads Abstract | Research in cellular mechanotransduction often focuses on how in living cells because their structural integrity extracellular physical forces are converted into chemical signals at the cell surface. is maintained for longer than the turnover However, mechanical forces that are exerted on surface-adhesion receptors, such time of individual protein monomers15–17. as integrins and cadherins, are also channelled along cytoskeletal filaments and Key to the cellular tensegrity model is concentrated at distant sites in the cytoplasm and nucleus. Here, we explore the the idea that overall cell-shape stability and long-distance force transfer are governed by molecular mechanisms by which forces might act at a distance to induce the level of isometric tension, or ‘prestress’, mechanochemical conversion in the nucleus and alter gene activities. in the cytoskeleton that is generated through the establishment of a force balance between Mechanical forces influence the growth and For example, endothelial cells sense fluid opposing structural elements (that is, micro- shape of virtually every tissue and organ in shear through a cell–cell junctional com- tubules, contractile microfilaments and our bodies. -

Supplemental Table 1
Supplemental Data Supplemental Table 1. Genes differentially regulated by Ad-KLF2 vs. Ad-GFP infected EC. Three independent genome-wide transcriptional profiling experiments were performed, and significantly regulated genes were identified. Color-coding scheme: Up, p < 1e-15 Up, 1e-15 < p < 5e-10 Up, 5e-10 < p < 5e-5 Up, 5e-5 < p <.05 Down, p < 1e-15 As determined by Zpool Down, 1e-15 < p < 5e-10 Down, 5e-10 < p < 5e-5 Down, 5e-5 < p <.05 p<.05 as determined by Iterative Standard Deviation Algorithm as described in Supplemental Methods Ratio RefSeq Number Gene Name 1,058.52 KRT13 - keratin 13 565.72 NM_007117.1 TRH - thyrotropin-releasing hormone 244.04 NM_001878.2 CRABP2 - cellular retinoic acid binding protein 2 118.90 NM_013279.1 C11orf9 - chromosome 11 open reading frame 9 109.68 NM_000517.3 HBA2;HBA1 - hemoglobin, alpha 2;hemoglobin, alpha 1 102.04 NM_001823.3 CKB - creatine kinase, brain 96.23 LYNX1 95.53 NM_002514.2 NOV - nephroblastoma overexpressed gene 75.82 CeleraFN113625 FLJ45224;PTGDS - FLJ45224 protein;prostaglandin D2 synthase 21kDa 74.73 NM_000954.5 (brain) 68.53 NM_205545.1 UNQ430 - RGTR430 66.89 NM_005980.2 S100P - S100 calcium binding protein P 64.39 NM_153370.1 PI16 - protease inhibitor 16 58.24 NM_031918.1 KLF16 - Kruppel-like factor 16 46.45 NM_024409.1 NPPC - natriuretic peptide precursor C 45.48 NM_032470.2 TNXB - tenascin XB 34.92 NM_001264.2 CDSN - corneodesmosin 33.86 NM_017671.3 C20orf42 - chromosome 20 open reading frame 42 33.76 NM_024829.3 FLJ22662 - hypothetical protein FLJ22662 32.10 NM_003283.3 TNNT1 - troponin T1, skeletal, slow LOC388888 (LOC388888), mRNA according to UniGene - potential 31.45 AK095686.1 CONFLICT - LOC388888 (na) according to LocusLink. -

Network of Micrornas-Mrnas Interactions in Pancreatic Cancer
Hindawi Publishing Corporation BioMed Research International Volume 2014, Article ID 534821, 8 pages http://dx.doi.org/10.1155/2014/534821 Research Article Network of microRNAs-mRNAs Interactions in Pancreatic Cancer Elnaz Naderi,1 Mehdi Mostafaei,2 Akram Pourshams,1 and Ashraf Mohamadkhani1 1 Liver and Pancreatobiliary Diseases Research Center, Digestive Diseases Research Institute, Tehran University of Medical Sciences, Tehran, Iran 2 Biotechnology Engineering, Islamic Azad University,Tehran North Branch, Tehran, Iran Correspondence should be addressed to Ashraf Mohamadkhani; [email protected] Received 5 February 2014; Revised 13 April 2014; Accepted 13 April 2014; Published 7 May 2014 Academic Editor: FangXiang Wu Copyright © 2014 Elnaz Naderi et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Background. MicroRNAs are small RNA molecules that regulate the expression of certain genes through interaction with mRNA targets and are mainly involved in human cancer. This study was conducted to make the network of miRNAs-mRNAs interactions in pancreatic cancer as the fourth leading cause of cancer death. Methods. 56 miRNAs that were exclusively expressed and 1176 genes that were downregulated or silenced in pancreas cancer were extracted from beforehand investigations. MiRNA–mRNA interactions data analysis and related networks were explored using MAGIA tool and Cytoscape 3 software. Functional annotations of candidate genes in pancreatic cancer were identified by DAVID annotation tool. Results. This network is made of 217 nodes for mRNA, 15 nodes for miRNA, and 241 edges that show 241 regulations between 15 miRNAs and 217 target genes. -

Genotype–Phenotype Correlations to Aid in the Prognosis Of
European Journal of Human Genetics (2007) 15, 446–452 & 2007 Nature Publishing Group All rights reserved 1018-4813/07 $30.00 www.nature.com/ejhg ARTICLE Genotype–phenotype correlations to aid in the prognosis of individuals with uncommon 20q13.33 subtelomere deletions: a collaborative study on behalf of the ‘association des Cytoge´ne´ticiens de langue Franc¸aise’ Myle`ne Be´ri-Deixheimer1, Marie-Jose´ Gregoire1, Annick Toutain2, Kare`ne Brochet1, Sylvain Briault2, Jean-Luc Schaff3, Bruno Leheup4 and Philippe Jonveaux*,1 1Laboratoire de Ge´ne´tique, EA 4002, CHU, Nancy-University, France; 2Service de Ge´ne´tique, Hoˆpital Bretonneau, Tours, France; 3Service de neurologie, CHU, Nancy-Univeristy, France; 4Service de me´decine infantile et ge´ne´tique clinique, CHU, Nancy-Univeristy, France The identification of subtelomeric rearrangements as a cause of mental retardation has made a considerable contribution to diagnosing patients with mental retardation. It is remarkable that for certain subtelomeric regions, deletions have hardly ever been reported so far. All the laboratories from the ‘Association des Cytoge´ne´ticiens de Langue Franc¸aise’ were surveyed for cases where an abnormality of the subtelomere FISH analysis had been ascertained. Among 1511 cases referred owing to unexplained mental retardation, 115 (7.6%) patients showed a clinically significant subtelomeric abnormality. We report the clinical features and the molecular cytogenetic delineation of isolated de novo deletions on 20q13.33 in two cases. Detailed mapping was performed by micro-array CGH in one patient and confirmed by FISH in the two patients. We compare our data with the only three patients reported in the literature. -

The SUN Rises on Meiotic Chromosome Dynamics
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Developmental Cell Review Feature The SUN Rises on Meiotic Chromosome Dynamics Yasushi Hiraoka1,2,* and Abby F. Dernburg3,4,* 1Graduate School of Frontier Biosciences, Osaka University, 1-3 Yamadaoka, Suita 565-0871, Japan 2Kobe Advanced ICT Research Center, National Institute of Information and Communications Technology, 588-2 Iwaoka, Iwaoka-cho, Nishi-ku, Kobe 651-2492, Japan 3Howard Hughes Medical Institute and Department of Molecular and Cell Biology, University of California, Berkeley, Berkeley, CA 94720-3220, USA 4Life Sciences Division, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA *Correspondence: [email protected] (Y.H.), [email protected] (A.F.D.) DOI 10.1016/j.devcel.2009.10.014 Recent studies in diverse eukaryotes have implicated a family of nuclear envelope proteins containing SUN domains as key components of meiotic nuclear organization and chromosome dynamics. In many cases, these transmembrane proteins are also known to contribute to centrosome or spindle pole body function in mitotically dividing cells. During meiotic prophase, the apparent role of these SUN-domain proteins, together with their partner KASH-domain proteins, is to connect chromosomes through the intact nuclear envelope to force-generating mechanisms in the cytoplasm. A Family of SUN Proteins In this review, we use these existing names and also refer to these The SUN domain was named for homologous sequences shared genes and their orthologs in mouse and human genomes as Sun4 by Sad1 and UNC-84 proteins (Malone et al., 1999).