Supplemental Table 1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

1 Vitamins D2 and D3 Have Overlapping but Different Effects On
medRxiv preprint doi: https://doi.org/10.1101/2020.12.16.20247700; this version posted February 22, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. It is made available under a CC-BY 4.0 International license . Vitamins D2 and D3 have overlapping but different effects on human gene expression revealed through analysis of blood transcriptomes in a randomised double-blind placebo-controlled food-fortification trial Louise R. Durrant,1, 2, Giselda Buccaa,b, 1,3, Andrew Heskethb, 1, Carla Möller-Leveta, Laura Tripkovica, Huihai Wua, Kathryn H. Harta, John C. Mathersc, Ruan M. Elliotta, Susan A. Lanham-Newa & Colin P. Smitha,b, 3* a Department of Nutritional Sciences, School of Biosciences and Medicine, Faculty of Health and Medical Sciences, University of Surrey, Guildford, UK b School of Pharmacy and Biomolecular Sciences, University of Brighton, Brighton, UK c Human Nutrition Research Centre, Population Health Sciences Institute, Newcastle University, UK 1 These authors contributed equally 2 Current address: Yakult UK Ltd., Anteros, Odyssey Business Park, West End Road, South Ruislip, HA4 6QQ; formerly known as Louise Wilson. 3 Current address: School of Pharmacy and Biomolecular Sciences, University of Brighton, Brighton, UK *For correspondence: Prof Colin P. Smith, School of Pharmacy and Biomolecular Sciences, University of Brighton, Huxley Building, Moulsecoomb, Brighton, BN2 4GJ, UK; Tel. +44 (0)1273 642048. Email: [email protected] Key words: Vitamin D3, vitamin D2, 25-hydroxyvitamin D, human transcriptome, immunity, ethnic differences, Covid-19 Abstract For the first time, we report the influence of vitamin D2 and vitamin D3 on genome- wide gene expression in whole blood from healthy women representing two ethnic groups, white European and South Asian. -

Differential Regulation of Gene Expression by Cholesterol Biosynthesis Inhibitors That Reduce (Pravastatin) Or Enhance (Squalest
JPET Fast Forward. Published on May 25, 2016 as DOI: 10.1124/jpet.116.233312 This article has not been copyedited and formatted. The final version may differ from this version. Differential regulation of gene expression by cholesterol biosynthesis inhibitors that reduce (pravastatin) or enhance (squalestatin 1) nonsterol isoprenoid levels in primary cultured mouse and rat hepatocytes. Elizabeth A. Rondini, Zofia Duniec-Dmuchowski, Daniela Cukovic, Alan A. Dombkowski, and Thomas A. Kocarek Downloaded from Institute of Environmental Health Sciences, Wayne State University, Detroit, MI 48202, USA (E.A.R., Z.D-D, T.A.K.) jpet.aspetjournals.org Department of Pediatrics, Division of Clinical Pharmacology and Toxicology, Wayne State University, Detroit, MI 48202 (D.C., A.A.D) at ASPET Journals on September 27, 2021 JPET Fast Forward. Published on May 25, 2016 as DOI: 10.1124/jpet.116.233312 This article has not been copyedited and formatted. The final version may differ from this version. JPET #233312 Running title: Regulation of hepatocellular gene expression by isoprenoids Address correspondence to: Dr. Thomas A. Kocarek, Institute of Environmental Health Sciences, 6135 Woodward Avenue, IBio Building, Room 2126, Wayne State University, Detroit, MI 48202, USA. Tel: (313) 577-6580; FAX: (313) 972-8025; E-mail: [email protected] Number of text pages: 43 Downloaded from Number of tables: 2 Supplemental Number of figures: 8 jpet.aspetjournals.org Number of references: 77 Number of words in Abstract: 249 Number of words in Introduction: 745 at -

RNA Expression Patterns in Serum Microvesicles from Patients With
Noerholm et al. BMC Cancer 2012, 12:22 http://www.biomedcentral.com/1471-2407/12/22 RESEARCHARTICLE Open Access RNA expression patterns in serum microvesicles from patients with glioblastoma multiforme and controls Mikkel Noerholm1,2*, Leonora Balaj1, Tobias Limperg1,3, Afshin Salehi1,4, Lin Dan Zhu1, Fred H Hochberg1, Xandra O Breakefield1, Bob S Carter1,4 and Johan Skog1,2 Abstract Background: RNA from exosomes and other microvesicles contain transcripts of tumour origin. In this study we sought to identify biomarkers of glioblastoma multiforme in microvesicle RNA from serum of affected patients. Methods: Microvesicle RNA from serum from patients with de-novo primary glioblastoma multiforme (N = 9) and normal controls (N = 7) were analyzed by microarray analysis. Samples were collected according to protocols approved by the Institutional Review Board. Differential expressions were validated by qRT-PCR in a separate set of samples (N = 10 in both groups). Results: Expression profiles of microvesicle RNA correctly separated individuals in two groups by unsupervised clustering. The most significant differences pertained to down-regulated genes (121 genes > 2-fold down) in the glioblastoma multiforme patient microvesicle RNA, validated by qRT-PCR on several genes. Overall, yields of microvesicle RNA from patients was higher than from normal controls, but the additional RNA was primarily of size < 500 nt. Gene ontology of the down-regulated genes indicated these are coding for ribosomal proteins and genes related to ribosome production. Conclusions: Serum microvesicle RNA from patients with glioblastoma multiforme has significantly down- regulated levels of RNAs coding for ribosome production, compared to normal healthy controls, but a large overabundance of RNA of unknown origin with size < 500 nt. -

Establishing the Pathogenicity of Novel Mitochondrial DNA Sequence Variations: a Cell and Molecular Biology Approach
Mafalda Rita Avó Bacalhau Establishing the Pathogenicity of Novel Mitochondrial DNA Sequence Variations: a Cell and Molecular Biology Approach Tese de doutoramento do Programa de Doutoramento em Ciências da Saúde, ramo de Ciências Biomédicas, orientada pela Professora Doutora Maria Manuela Monteiro Grazina e co-orientada pelo Professor Doutor Henrique Manuel Paixão dos Santos Girão e pela Professora Doutora Lee-Jun C. Wong e apresentada à Faculdade de Medicina da Universidade de Coimbra Julho 2017 Faculty of Medicine Establishing the pathogenicity of novel mitochondrial DNA sequence variations: a cell and molecular biology approach Mafalda Rita Avó Bacalhau Tese de doutoramento do programa em Ciências da Saúde, ramo de Ciências Biomédicas, realizada sob a orientação científica da Professora Doutora Maria Manuela Monteiro Grazina; e co-orientação do Professor Doutor Henrique Manuel Paixão dos Santos Girão e da Professora Doutora Lee-Jun C. Wong, apresentada à Faculdade de Medicina da Universidade de Coimbra. Julho, 2017 Copyright© Mafalda Bacalhau e Manuela Grazina, 2017 Esta cópia da tese é fornecida na condição de que quem a consulta reconhece que os direitos de autor são pertença do autor da tese e do orientador científico e que nenhuma citação ou informação obtida a partir dela pode ser publicada sem a referência apropriada e autorização. This copy of the thesis has been supplied on the condition that anyone who consults it recognizes that its copyright belongs to its author and scientific supervisor and that no quotation from the -

Identification and Characterization of Zebrafish 17Beta-HSD Type 1 and Type 3: a Comparative Analysis of Androgen/Estrogen Activity Regulators
Institut für Experimentelle Genetik GSF-Forschungzentrum für Umwelt und Gesundheit, Neuherberg Identification and characterization of zebrafish 17beta-HSD type 1 and type 3: A comparative analysis of androgen/estrogen activity regulators Rebekka Mindnich Vollständiger Abdruck der von der Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt der Technischen Universität München zur Erlangung des akademischen Grades eines Doktors der Naturwissenschaften genehmigten Dissertation. Vorsitzender: Univ.- Prof. Dr. Bertold Hock Prüfer der Dissertation: 1. Priv.-Doz. Dr. Jerzy Adamski 2. Univ.-Prof. Dr. Johannes Buchner 3. Univ.-Prof. Dr. Wolfgang Wurst Die Dissertation wurde am 30.06.2004 bei der Technischen Universität München eingereicht und durch die Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt am 07.10. 2004 angenommen. Table of contents Table of contents ABSTRACT................................................................................................................................... 7 ZUSAMMENFASSUNG................................................................................................................ 9 ABBREVIATIONS....................................................................................................................... 11 1 INTRODUCTION ................................................................................................................ 13 1.1 THE AIM OF THIS STUDY ............................................................................................... -

Frontiers in Integrative Genomics and Translational Bioinformatics
BioMed Research International Frontiers in Integrative Genomics and Translational Bioinformatics Guest Editors: Zhongming Zhao, Victor X. Jin, Yufei Huang, Chittibabu Guda, and Jianhua Ruan Frontiers in Integrative Genomics and Translational Bioinformatics BioMed Research International Frontiers in Integrative Genomics and Translational Bioinformatics Guest Editors: Zhongming Zhao, Victor X. Jin, Yufei Huang, Chittibabu Guda, and Jianhua Ruan Copyright © òýÔ Hindawi Publishing Corporation. All rights reserved. is is a special issue published in “BioMed Research International.” All articles are open access articles distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Contents Frontiers in Integrative Genomics and Translational Bioinformatics, Zhongming Zhao, Victor X. Jin, Yufei Huang, Chittibabu Guda, and Jianhua Ruan Volume òýÔ , Article ID Þò ¥ÀÔ, ç pages Building Integrated Ontological Knowledge Structures with Ecient Approximation Algorithms, Yang Xiang and Sarath Chandra Janga Volume òýÔ , Article ID ýÔ ò, Ô¥ pages Predicting Drug-Target Interactions via Within-Score and Between-Score, Jian-Yu Shi, Zun Liu, Hui Yu, and Yong-Jun Li Volume òýÔ , Article ID ç ýÀç, À pages RNAseq by Total RNA Library Identies Additional RNAs Compared to Poly(A) RNA Library, Yan Guo, Shilin Zhao, Quanhu Sheng, Mingsheng Guo, Brian Lehmann, Jennifer Pietenpol, David C. Samuels, and Yu Shyr Volume òýÔ , Article ID âòÔçý, À pages Construction of Pancreatic Cancer Classier Based on SVM Optimized by Improved FOA, Huiyan Jiang, Di Zhao, Ruiping Zheng, and Xiaoqi Ma Volume òýÔ , Article ID ÞÔýòç, Ôò pages OperomeDB: A Database of Condition-Specic Transcription Units in Prokaryotic Genomes, Kashish Chetal and Sarath Chandra Janga Volume òýÔ , Article ID çÔòÔÞ, Ôý pages How to Choose In Vitro Systems to Predict In Vivo Drug Clearance: A System Pharmacology Perspective, Lei Wang, ChienWei Chiang, Hong Liang, Hengyi Wu, Weixing Feng, Sara K. -

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

Meta-Analysis of Nasopharyngeal Carcinoma
BMC Genomics BioMed Central Research article Open Access Meta-analysis of nasopharyngeal carcinoma microarray data explores mechanism of EBV-regulated neoplastic transformation Xia Chen†1,2, Shuang Liang†1, WenLing Zheng1,3, ZhiJun Liao1, Tao Shang1 and WenLi Ma*1 Address: 1Institute of Genetic Engineering, Southern Medical University, Guangzhou, PR China, 2Xiangya Pingkuang associated hospital, Pingxiang, Jiangxi, PR China and 3Southern Genomics Research Center, Guangzhou, Guangdong, PR China Email: Xia Chen - [email protected]; Shuang Liang - [email protected]; WenLing Zheng - [email protected]; ZhiJun Liao - [email protected]; Tao Shang - [email protected]; WenLi Ma* - [email protected] * Corresponding author †Equal contributors Published: 7 July 2008 Received: 16 February 2008 Accepted: 7 July 2008 BMC Genomics 2008, 9:322 doi:10.1186/1471-2164-9-322 This article is available from: http://www.biomedcentral.com/1471-2164/9/322 © 2008 Chen et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: Epstein-Barr virus (EBV) presumably plays an important role in the pathogenesis of nasopharyngeal carcinoma (NPC), but the molecular mechanism of EBV-dependent neoplastic transformation is not well understood. The combination of bioinformatics with evidences from biological experiments paved a new way to gain more insights into the molecular mechanism of cancer. Results: We profiled gene expression using a meta-analysis approach. Two sets of meta-genes were obtained. Meta-A genes were identified by finding those commonly activated/deactivated upon EBV infection/reactivation. -

Figure S1. DMD Module Network. the Network Is Formed by 260 Genes from Disgenet and 1101 Interactions from STRING. Red Nodes Are the Five Seed Candidate Genes
Figure S1. DMD module network. The network is formed by 260 genes from DisGeNET and 1101 interactions from STRING. Red nodes are the five seed candidate genes. Figure S2. DMD module network is more connected than a random module of the same size. It is shown the distribution of the largest connected component of 10.000 random modules of the same size of the DMD module network. The green line (x=260) represents the DMD largest connected component, obtaining a z-score=8.9. Figure S3. Shared genes between BMD and DMD signature. A) A meta-analysis of three microarray datasets (GSE3307, GSE13608 and GSE109178) was performed for the identification of differentially expressed genes (DEGs) in BMD muscle biopsies as compared to normal muscle biopsies. Briefly, the GSE13608 dataset included 6 samples of skeletal muscle biopsy from healthy people and 5 samples from BMD patients. Biopsies were taken from either biceps brachii, triceps brachii or deltoid. The GSE3307 dataset included 17 samples of skeletal muscle biopsy from healthy people and 10 samples from BMD patients. The GSE109178 dataset included 14 samples of controls and 11 samples from BMD patients. For both GSE3307 and GSE10917 datasets, biopsies were taken at the time of diagnosis and from the vastus lateralis. For the meta-analysis of GSE13608, GSE3307 and GSE109178, a random effects model of effect size measure was used to integrate gene expression patterns from the two datasets. Genes with an adjusted p value (FDR) < 0.05 and an │effect size│>2 were identified as DEGs and selected for further analysis. A significant number of DEGs (p<0.001) were in common with the DMD signature genes (blue nodes), as determined by a hypergeometric test assessing the significance of the overlap between the BMD DEGs and the number of DMD signature genes B) MCODE analysis of the overlapping genes between BMD DEGs and DMD signature genes. -

Download (PDF)
Tomono et al.: Glycan evolution based on phylogenetic profiling 1 Supplementary Table S1. List of 173 enzymes that are composed of glycosyltransferases and functionally-linked glycan synthetic enzymes UniProt ID Protein Name Categories of Glycan Localization CAZy Class 1 Q8N5D6 Globoside -1,3-N -acetylgalactosaminyltransferase 1 Glycosphingolipid Golgi apparatus GT6 P16442 Histo-blood group ABO system transferase Glycosphingolipid Golgi apparatus GT6 P19526 Galactoside 2--L-fucosyltransferase 1 Glycosphingolipid Golgi apparatus GT11 Q10981 Galactoside 2--L-fucosyltransferase 2 Glycosphingolipid Golgi apparatus GT11 Q00973 -1,4 N -acetylgalactosaminyltransferase 1 Glycosphingolipid Golgi apparatus GT12 Q8NHY0 -1,4 N -acetylgalactosaminyltransferase 2 O -Glycan, N -Glycan, Glycosphingolipid Golgi apparatus GT12 Q09327 -1,4-mannosyl-glycoprotein 4--N -acetylglucosaminyltransferase N -Glycan Golgi apparatus GT17 Q09328 -1,6-mannosylglycoprotein 6--N -acetylglucosaminyltransferase A N -Glycan Golgi apparatus GT18 Q3V5L5 -1,6-mannosylglycoprotein 6--N -acetylglucosaminyltransferase B O -Glycan, N -Glycan Golgi apparatus GT18 Q92186 -2,8-sialyltransferase 8B (SIAT8-B) (ST8SiaII) (STX) N -Glycan Golgi apparatus GT29 O15466 -2,8-sialyltransferase 8E (SIAT8-E) (ST8SiaV) Glycosphingolipid Golgi apparatus GT29 P61647 -2,8-sialyltransferase 8F (SIAT8-F) (ST8SiaVI) O -Glycan Golgi apparatus GT29 Q9NSC7 -N -acetylgalactosaminide -2,6-sialyltransferase 1 (ST6GalNAcI) (SIAT7-A) O -Glycan Golgi apparatus GT29 Q9UJ37 -N -acetylgalactosaminide -2,6-sialyltransferase -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -
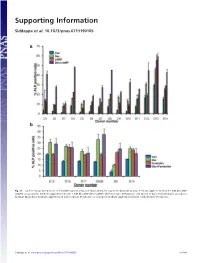
Supporting Information
Supporting Information Siddappa et al. 10.1073/pnas.0711190105 Fig. S1. (a) Percentage ALP-positive cells in hMSCs grown in basic medium (Con), osteogenic medium (Dex), basic medium supplemented with 1 mM db-cAMP (cAMP), or osteogenic medium supplemented with 1 mM db-cAMP (DexϩcAMP). (b) Percentage ALP-positive cells grown in basic medium (Con), osteogenic medium (Dex), basic medium supplemented with forskolin (Forskolin), or osteogenic medium supplemented with forskolin (DexϩForskolin). Siddappa et al. www.pnas.org/cgi/content/short/0711190105 1of10 Fig. S2. hMSCs were grown in basic medium, basic medium supplemented with 1 mM db-cAMP (cAMP), osteogenic medium (Dex), or osteogenic medium supplemented with 1 mM db-cAMP (DexϩcAMP). Expression was analyzed by qPCR and is expressed as fold induction compared with cells grown in basic medium. The data were analyzed by using two-way ANOVA, and statistical significance is indicated compared with cells grown in basic medium. *, P Ͻ 0.05. Siddappa et al. www.pnas.org/cgi/content/short/0711190105 2of10 Fig. S3. (a) Methylene blue staining of hMSC-seeded scaffolds grown in basic medium (Con) or basic medium supplemented with 1 mM db-cAMP (cAMP) for 4 days. Note the less intensely stained db-cAMP-treated construct, indicating reduced cell numbers. (b) Quantitative Alamar blue assay for cell number analysis. The data were analyzed by using one-way ANOVA followed by Dunnet’s multiple-comparison test. Statistical significance is indicated compared with cells grown in basic medium (Con). *, P Ͻ 0.05 Siddappa et al. www.pnas.org/cgi/content/short/0711190105 3of10 Fig.