Decision Summary
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Legionella Shows a Diverse Secondary Metabolism Dependent on a Broad Spectrum Sfp-Type Phosphopantetheinyl Transferase
Legionella shows a diverse secondary metabolism dependent on a broad spectrum Sfp-type phosphopantetheinyl transferase Nicholas J. Tobias1, Tilman Ahrendt1, Ursula Schell2, Melissa Miltenberger1, Hubert Hilbi2,3 and Helge B. Bode1,4 1 Fachbereich Biowissenschaften, Merck Stiftungsprofessur fu¨r Molekulare Biotechnologie, Goethe Universita¨t, Frankfurt am Main, Germany 2 Max von Pettenkofer Institute, Ludwig-Maximilians-Universita¨tMu¨nchen, Munich, Germany 3 Institute of Medical Microbiology, University of Zu¨rich, Zu¨rich, Switzerland 4 Buchmann Institute for Molecular Life Sciences, Goethe Universita¨t, Frankfurt am Main, Germany ABSTRACT Several members of the genus Legionella cause Legionnaires’ disease, a potentially debilitating form of pneumonia. Studies frequently focus on the abundant number of virulence factors present in this genus. However, what is often overlooked is the role of secondary metabolites from Legionella. Following whole genome sequencing, we assembled and annotated the Legionella parisiensis DSM 19216 genome. Together with 14 other members of the Legionella, we performed comparative genomics and analysed the secondary metabolite potential of each strain. We found that Legionella contains a huge variety of biosynthetic gene clusters (BGCs) that are potentially making a significant number of novel natural products with undefined function. Surprisingly, only a single Sfp-like phosphopantetheinyl transferase is found in all Legionella strains analyzed that might be responsible for the activation of all carrier proteins in primary (fatty acid biosynthesis) and secondary metabolism (polyketide and non-ribosomal peptide synthesis). Using conserved active site motifs, we predict Submitted 29 June 2016 some novel compounds that are probably involved in cell-cell communication, Accepted 25 October 2016 Published 24 November 2016 differing to known communication systems. -

The Risk to Human Health from Free-Living Amoebae Interaction with Legionella in Drinking and Recycled Water Systems
THE RISK TO HUMAN HEALTH FROM FREE-LIVING AMOEBAE INTERACTION WITH LEGIONELLA IN DRINKING AND RECYCLED WATER SYSTEMS Dissertation submitted by JACQUELINE MARIE THOMAS BACHELOR OF SCIENCE (HONOURS) AND BACHELOR OF ARTS, UNSW In partial fulfillment of the requirements for the award of DOCTOR OF PHILOSOPHY in ENVIRONMENTAL ENGINEERING SCHOOL OF CIVIL AND ENVIRONMENTAL ENGINEERING FACULTY OF ENGINEERING MAY 2012 SUPERVISORS Professor Nicholas Ashbolt Office of Research and Development United States Environmental Protection Agency Cincinnati, Ohio USA and School of Civil and Environmental Engineering Faculty of Engineering The University of New South Wales Sydney, Australia Professor Richard Stuetz School of Civil and Environmental Engineering Faculty of Engineering The University of New South Wales Sydney, Australia Doctor Torsten Thomas School of Biotechnology and Biomolecular Sciences Faculty of Science The University of New South Wales Sydney, Australia ORIGINALITY STATEMENT '1 hereby declare that this submission is my own work and to the best of my knowledge it contains no materials previously published or written by another person, or substantial proportions of material which have been accepted for the award of any other degree or diploma at UNSW or any other educational institution, except where due acknowledgement is made in the thesis. Any contribution made to the research by others, with whom 1 have worked at UNSW or elsewhere, is explicitly acknowledged in the thesis. I also declare that the intellectual content of this thesis is the product of my own work, except to the extent that assistance from others in the project's design and conception or in style, presentation and linguistic expression is acknowledged.' Signed ~ ............................ -

Electronic Laboratory Reporting Use Case January 2011
ILLINOIS HEALTH INFORMATION EXCHANGE Electronic Laboratory Reporting Use Case Electronic Laboratory Reporting and Health Information Exchange Illinois Health Information Exchange Public Health Work Group January 2011 Electronic Laboratory Reporting Use Case January 2011 Table of Contents 1.0 Executive Summary……………………………………………………………………….3 2.0 Introduction…………………………………………………………………...……………..5 3.0 Scope……………………………………..………………………………………………………5 4.0 Use Case Stakeholders…………………………………………………………….….....6 5.0 Issues and Obstacles……………………………………………………………………...8 6.0 Use Case Pre-Conditions .………………….…………………………………………...8 7.0 Use Case Post-Conditions.……………………………………………………………...9 8.0 Detailed Scenarios/Technical Specifications.………………………………10 9.0 Validation and Certification………………………………………………………...12 Appendix ………………………………………………………………………………………….....13 Page 2 Electronic Laboratory Reporting Use Case January 2011 1.0 Executive Summary This Use Case is a product of the Public Health Work Group (PHWG) of the Illinois Health Information Exchange (HIE) Advisory Committee. The Illinois HIE Advisory Committee was constituted as the diverse public healthcare stakeholder body providing input and recommendations on the creation of the Illinois Health Information Exchange Authority (“the Authority”) as the Illinois vehicle for designing and implementing electronic health information exchange in Illinois. The establishment of the Authority marks the formal transition of the work of the HIE Advisory Committee and the Work Groups into alignment with the provisions of Illinois -
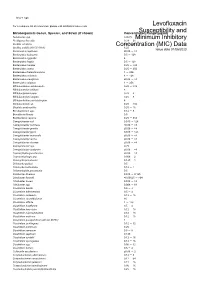
Susceptibility and Resistance Data
toku-e logo For a complete list of references, please visit antibiotics.toku-e.com Levofloxacin Microorganism Genus, Species, and Strain (if shown) Concentration Range (μg/ml)Susceptibility and Aeromonas spp. 0.0625 Minimum Inhibitory Alcaligenes faecalis 0.39 - 25 Bacillus circulans Concentration0.25 - 8 (MIC) Data Bacillus subtilis (ATCC 6051) 6.25 Issue date 01/06/2020 Bacteroides capillosus ≤0.06 - >8 Bacteroides distasonis 0.5 - 128 Bacteroides eggerthii 4 Bacteroides fragilis 0.5 - 128 Bacteroides merdae 0.25 - >32 Bacteroides ovatus 0.25 - 256 Bacteroides thetaiotaomicron 1 - 256 Bacteroides uniformis 4 - 128 Bacteroides ureolyticus ≤0.06 - >8 Bacteroides vulgatus 1 - 256 Bifidobacterium adolescentis 0.25 - >32 Bifidobacterium bifidum 8 Bifidobacterium breve 0.25 - 8 Bifidobacterium longum 0.25 - 8 Bifidobacterium pseudolongum 8 Bifidobacterium sp. 0.25 - >32 Bilophila wadsworthia 0.25 - 16 Brevibacterium spp. 0.12 - 8 Brucella melitensis 0.5 Burkholderia cepacia 0.25 - 512 Campylobacter coli 0.015 - 128 Campylobacter concisus ≤0.06 - >8 Campylobacter gracilis ≤0.06 - >8 Campylobacter jejuni 0.015 - 128 Campylobacter mucosalis ≤0.06 - >8 Campylobacter rectus ≤0.06 - >8 Campylobacter showae ≤0.06 - >8 Campylobacter spp. 0.25 Campylobacter sputorum ≤0.06 - >8 Capnocytophaga ochracea ≤0.06 - >8 Capnocytophaga spp. 0.006 - 2 Chlamydia pneumonia 0.125 - 1 Chlamydia psittaci 0.5 Chlamydia trachomatis 0.12 - 1 Chlamydophila pneumonia 0.5 Citrobacter diversus 0.015 - 0.125 Citrobacter freundii ≤0.00625 - >64 Citrobacter koseri 0.015 - -

Aquascreen® Legionella Species Qpcr Detection Kit
AquaScreen® Legionella species qPCR Detection Kit INSTRUCTIONS FOR USE FOR USE IN RESEARCH AND QUALITY CONTROL Symbols Lot No. Cat. No. Expiry date Storage temperature Number of reactions Manufacturer INDICATION The AquaScreen® Legionella species qPCR Detection kit is specifically designed for the quantitative detection of several Legionella species in water samples prepared with the AquaScreen® FastExt- ract kit. Its design complies with the requirements of AFNOR T90-471 and ISO/TS 12869:2012. Legionella are ubiquitous bacteria in surface water and moist soil, where they parasitize protozoa. The optimal growth temperature lies between +15 and +45 °C, whereas these gram-negative bacteria are dormant below 20 °C and do not survive above 60 °C. Importantly, Legionella are well-known as opportunistic intracellular human pathogens causing Legionnaires’ disease and Pontiac fever. The transmission occurs through inhalation of contami- nated aerosols generated by an infected source (e.g. human-made water systems like shower- heads, sink faucets, heaters, cooling towers, and many more). In order to efficiently prevent Legionella outbreaks, water safety control measures need syste- matic application but also reliable validation by fast Legionella testing. TEST PRINCIPLE The AquaScreen® Legionella species Kit uses qPCR for quantitative detection of legionella in wa- ter samples. In contrast to more time-consuming culture-based methods, AquaScreen® assays need less than six hours including sample preparation and qPCR to reliably detect Legionella. Moreover, the AquaScreen® qPCR assay has proven excellent performance in terms of specificity and sensitivity: other bacterial genera remain undetected whereas linear quantification is obtai- ned up to 1 x 106 particles per sample, therefore requiring no material dilution. -

Immunoproteomic Identification of Biomarkers for Diagnosis of Legionellosis
Immunoproteomic identification of biomarkers for diagnosis of legionellosis Submitted in total fulfilment of the requirements for the degree of Doctor of philosophy by Kaylass Poorun Department of Chemistry and Biotechnology Faculty of Science, Engineering and Technology Swinburne University of Technology Australia 2014 Abstract Abstract Legionellosis, a disease with significant mortality and morbidity rates, is considered to be the second most frequent cause of severe community-acquired pneumonia. It is difficult to distinguish from other types of pneumonia due to similar clinical manifestations. Several studies have demonstrated the inadequacies of current diagnostic tests for confirming Legionella infections. This study was aimed at identifying biomarkers that can be used in an improved test. A comparative proteomic analysis, using DIGE, was carried out between L. pneumophila ATCC33152 and L. longbeachae NSW150 and D4968 isolates. While many homologous proteins were found to be commonly expressed, numerous others were identified to be differentially expressed under similar in vitro conditions suggesting that the two species have different lifestyles and infection strategies. The bacterial immunoglobulin domain containing protein, found to share sequence homology to Type V secretion proteins intimin and invasin, is not known to be present in Legionella. Human sera containing antibodies against Legionella from a set of blind samples were identified by ELISA. Downstream analyses revealed that diverse immunogens may be responsible for eliciting immune response in different Legionella species which in turn show little to no congeneric cross-reactivity. To the best of our knowledge, this is a unique finding not previously reported. Several serological diagnostic tests currently in use do not include many Legionella species in their testing panel, which may be a reason for many Legionella species being under-reported. -

APUTS) Reporting Terminology and Codes Microbiology (V1.0
AUSTRALIAN PATHOLOGY UNITS AND TERMINOLOGY (APUTS) Reporting Terminology and Codes Microbiology (v1.0) 1 12/02/2013 APUTS Report Information Model - Urine Microbiology Page 1 of 1 Specimen Type Specimen Macro Time Glucose Bilirubin Ketones Specific Gravity pH Chemistry Protein Urobilinogen Nitrites Haemoglobin Leucocyte Esterases White blood cell count Red blood cells Cells Epithelial cells Bacteria Microscopy Parasites Microorganisms Yeasts Casts Crystals Other elements Antibacterial Activity No growth Mixed growth Urine MCS No significant growth Klebsiella sp. Bacteria ESBL Klebsiella pneumoniae Identification Virus Fungi Growth of >10^8 org/L 10^7 to 10^8 organism/L of mixed Range or number Colony Count growth of 3 organisms 19090-0 Culture Organism 1 630-4 LOINC >10^8 organisms/L LOINC Significant growth e.g. Ampicillin 18864-9 LOINC Antibiotics Susceptibility Method Released/suppressed None Organism 2 Organism 3 Organism 4 None Consistent with UTI Probable contamination Growth unlikely to be significant Comment Please submit a repeat specimen for testing if clinically indicated Catheter comments Sterile pyuria Notification to infection control and public health departments PUTS Urine Microbiology Information Model v1.mmap - 12/02/2013 - Mindjet 12/02/2013 APUTS Report Terminology and Codes - Microbiology - Urine Page 1 of 3 RCPA Pathology Units and Terminology Standardisation Project - Terminology for Reporting Pathology: Microbiology : Urine Microbiology Report v1 LOINC LOINC LOINC LOINC LOINC LOINC LOINC Urine Microbiology Report -

CGM-18-001 Perseus Report Update Bacterial Taxonomy Final Errata
report Update of the bacterial taxonomy in the classification lists of COGEM July 2018 COGEM Report CGM 2018-04 Patrick L.J. RÜDELSHEIM & Pascale VAN ROOIJ PERSEUS BVBA Ordering information COGEM report No CGM 2018-04 E-mail: [email protected] Phone: +31-30-274 2777 Postal address: Netherlands Commission on Genetic Modification (COGEM), P.O. Box 578, 3720 AN Bilthoven, The Netherlands Internet Download as pdf-file: http://www.cogem.net → publications → research reports When ordering this report (free of charge), please mention title and number. Advisory Committee The authors gratefully acknowledge the members of the Advisory Committee for the valuable discussions and patience. Chair: Prof. dr. J.P.M. van Putten (Chair of the Medical Veterinary subcommittee of COGEM, Utrecht University) Members: Prof. dr. J.E. Degener (Member of the Medical Veterinary subcommittee of COGEM, University Medical Centre Groningen) Prof. dr. ir. J.D. van Elsas (Member of the Agriculture subcommittee of COGEM, University of Groningen) Dr. Lisette van der Knaap (COGEM-secretariat) Astrid Schulting (COGEM-secretariat) Disclaimer This report was commissioned by COGEM. The contents of this publication are the sole responsibility of the authors and may in no way be taken to represent the views of COGEM. Dit rapport is samengesteld in opdracht van de COGEM. De meningen die in het rapport worden weergegeven, zijn die van de auteurs en weerspiegelen niet noodzakelijkerwijs de mening van de COGEM. 2 | 24 Foreword COGEM advises the Dutch government on classifications of bacteria, and publishes listings of pathogenic and non-pathogenic bacteria that are updated regularly. These lists of bacteria originate from 2011, when COGEM petitioned a research project to evaluate the classifications of bacteria in the former GMO regulation and to supplement this list with bacteria that have been classified by other governmental organizations. -

WO 2012/055408 Al
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date . 3 May 2012 (03.05.2012) WO 2012/055408 Al (51) International Patent Classification: DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, CI2Q 1/68 (2006.01) HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KM, KN, KP, KR, KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, (21) International Application Number: ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, PCT/DK20 11/000120 NO, NZ, OM, PE, PG, PH, PL, PT, QA, RO, RS, RU, (22) International Filing Date: RW, SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, 27 October 201 1 (27.10.201 1) TM, TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every (26) Publication Language: English kind of regional protection available): ARIPO (BW, GH, (30) Priority Data: GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, SZ, TZ, 61/407,122 27 October 2010 (27.10.2010) US UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, MD, PA 2010 70455 27 October 2010 (27.10.2010) DK RU, TJ, TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, EE, ES, FI, FR, GB, GR, HR, HU, IE, IS, IT, (71) Applicant (for all designated States except US): QUAN- LT, LU, LV, MC, MK, MT, NL, NO, PL, PT, RO, RS, TIBACT A/S [DK/DK]; Kettegards Alle 30, DK-2650 SE, SI, SK, SM, TR), OAPI (BF, BJ, CF, CG, CI, CM, Hvidovre (DK). -

ESCMID Online Lecture Library © by Author ESCMID Online Lecture Library Latex Agglutination Test
The etiological agent: Legionella pneumophila and other Legionella spp. Valeria Gaia © by author National Reference Centre for Legionella ESCMIDc/o Online Microbiology Lecture laboratory Library Ente Ospedaliero Cantonale Bellinzona - Switzerland © by author ESCMID Online Lecture Library Hystory of Legionnaires’ Disease July 21st 1976 - Philadelphia • 58th Convention of the American Legion at the Bellevue-Stratford Hotel • > 4000 World War II Veterans with families & friends • 600 persons staying at the hotel © by author • ESCMIDJuly 23nd: convention Online closed Lecture Library • Several veterans showed symptoms of pneumonia Searching for the causative agent David Fraser: CDC – Atlanta •Influenza virus? •Nickel intoxication? •Toxin? o 2603 toxicology tests o 5120 microscopy exams o 990 serological tests© by author ESCMIDEverybody seems Online to agree: Lecture it’s NOT a bacterialLibrary disease! July 22nd – August 2nd •High fever •Coughing •Breathing difficulties •Chest pains •Exposed Population =© people by authorstaying in the lobby or outside the Bellevue Stratford Hotel «Broad Street Pneumonia» •221ESCMID persons were Online infected (182+39 Lecture «Broad StreetLibrary Pneumonia» ) 34 patients died (29+5) September 1976-January 1977 Joseph McDade: aims to rule out Q-fever (Rickettsiae) •Injection of “infected” pulmonary tissue in Guinea Pigs microscopy: Cocci and small Bacilli not significant at the time •Inoculation in embryonated eggs + antibiotics to inhibit the growth of contaminating bacteria No growth Microscopy on the -

Microbial Source Tracking in Coastal Recreational Waters of Southern Maine
University of New Hampshire University of New Hampshire Scholars' Repository Master's Theses and Capstones Student Scholarship Fall 2017 MICROBIAL SOURCE TRACKING IN COASTAL RECREATIONAL WATERS OF SOUTHERN MAINE: RELATIONSHIPS BETWEEN ENTEROCOCCI, ENVIRONMENTAL FACTORS, POTENTIAL PATHOGENS, AND FECAL SOURCES Derek Rothenheber University of New Hampshire, Durham Follow this and additional works at: https://scholars.unh.edu/thesis Recommended Citation Rothenheber, Derek, "MICROBIAL SOURCE TRACKING IN COASTAL RECREATIONAL WATERS OF SOUTHERN MAINE: RELATIONSHIPS BETWEEN ENTEROCOCCI, ENVIRONMENTAL FACTORS, POTENTIAL PATHOGENS, AND FECAL SOURCES" (2017). Master's Theses and Capstones. 1133. https://scholars.unh.edu/thesis/1133 This Thesis is brought to you for free and open access by the Student Scholarship at University of New Hampshire Scholars' Repository. It has been accepted for inclusion in Master's Theses and Capstones by an authorized administrator of University of New Hampshire Scholars' Repository. For more information, please contact [email protected]. MICROBIAL SOURCE TRACKING IN COASTAL RECREATIONAL WATERS OF SOUTHERN MAINE: RELATIONSHIPS BETWEEN ENTEROCOCCI, ENVIRONMENTAL FACTORS, POTENTIAL PATHOGENS, AND FECAL SOURCES BY DEREK ROTHENHEBER Microbiology BS, University of Maine, 2013 THESIS Submitted to the University of New Hampshire In Partial Fulfillment of the Requirements for the Degree of Master of Science in Microbiology September, 2017 This thesis has been examined and approved in partial fulfillment of the -

Legionella and Non-Tuberculous Mycobacteria Using MALDI TOF MS (Matrix Assisted Laser Desorption Ionisation Time of Flight Mass Spectrometry)
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by OTHES DIPLOMARBEIT Titel der Diplomarbeit Establishment of a reference database for Acanthamoeba, Legionella and non-tuberculous mycobacteria using MALDI TOF MS (Matrix Assisted Laser Desorption Ionisation Time of Flight Mass Spectrometry) Verfasserin Dzenita HASANACEVIC angestrebter akademischer Grad Magistra der Naturwissenschaften (Mag.rer.nat.) Wien, 2012 Studienkennzahl lt. A 442 Studienblatt: Studienrichtung lt. Studienblatt: Anthropologie Betreuerin: Ass. Prof. Univ. Doz. Mag. Dr. Julia Walochnik Contents 1 ABBREVIATIONS ..................................................................................................... 5 2 INTRODUCTION ....................................................................................................... 6 2.1 Acanthamoeba .................................................................................................... 6 2.1.1 Classification ................................................................................................ 6 2.1.1.1 Phylogeny of Acanthamoeba ................................................................. 6 2.1.1.2 Methods of classification ....................................................................... 8 2.1.2 Ecology and geographical distribution ........................................................ 11 2.1.2.1 Life cycle ............................................................................................. 11 2.1.2.2 Trophozoites ......................................................................................