Identification and Characterization of Differentially-Regulated Type Ivb
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Reporting of Diseases and Conditions Regulation, Amendment, M.R. 289/2014
THE PUBLIC HEALTH ACT LOI SUR LA SANTÉ PUBLIQUE (C.C.S.M. c. P210) (c. P210 de la C.P.L.M.) Reporting of Diseases and Conditions Règlement modifiant le Règlement sur la Regulation, amendment déclaration de maladies et d'affections Regulation 289/2014 Règlement 289/2014 Registered December 23, 2014 Date d'enregistrement : le 23 décembre 2014 Manitoba Regulation 37/2009 amended Modification du R.M. 37/2009 1 The Reporting of Diseases and 1 Le présent règlement modifie le Conditions Regulation , Manitoba Règlement sur la déclaration de maladies et Regulation 37/2009, is amended by this d'affections , R.M. 37/2009. regulation. 2 Schedules A and B are replaced with 2 Les annexes A et B sont remplacées Schedules A and B to this regulation. par les annexes A et B du présent règlement. Coming into force Entrée en vigueur 3 This regulation comes into force on 3 Le présent règlement entre en vigueur January 1, 2015, or on the day it is registered le 1 er janvier 2015 ou à la date de son under The Statutes and Regulations Act , enregistrement en vertu de Loi sur les textes whichever is later. législatifs et réglementaires , si cette date est postérieure. December 19, 2014 Minister of Health/La ministre de la Santé, 19 décembre 2014 Sharon Blady 1 SCHEDULE A (Section 1) 1 The following diseases are diseases requiring contact notification in accordance with the disease-specific protocol. Common name Scientific or technical name of disease or its infectious agent Chancroid Haemophilus ducreyi Chlamydia Chlamydia trachomatis (including Lymphogranuloma venereum (LGV) serovars) Gonorrhea Neisseria gonorrhoeae HIV Human immunodeficiency virus Syphilis Treponema pallidum subspecies pallidum Tuberculosis Mycobacterium tuberculosis Mycobacterium africanum Mycobacterium canetti Mycobacterium caprae Mycobacterium microti Mycobacterium pinnipedii Mycobacterium bovis (excluding M. -

Genomic Signatures of Predatory Bacteria
The ISME Journal (2013) 7, 756–769 & 2013 International Society for Microbial Ecology All rights reserved 1751-7362/13 www.nature.com/ismej ORIGINAL ARTICLE By their genes ye shall know them: genomic signatures of predatory bacteria Zohar Pasternak1, Shmuel Pietrokovski2, Or Rotem1, Uri Gophna3, Mor N Lurie-Weinberger3 and Edouard Jurkevitch1 1Department of Plant Pathology and Microbiology, The Hebrew University of Jerusalem, Rehovot, Israel; 2Department of Molecular Genetics, Weizmann Institute of Science, Rehovot, Israel and 3Department of Molecular Microbiology and Biotechnology, George S. Wise Faculty of Life Sciences, Tel Aviv University, Tel Aviv, Israel Predatory bacteria are taxonomically disparate, exhibit diverse predatory strategies and are widely distributed in varied environments. To date, their predatory phenotypes cannot be discerned in genome sequence data thereby limiting our understanding of bacterial predation, and of its impact in nature. Here, we define the ‘predatome,’ that is, sets of protein families that reflect the phenotypes of predatory bacteria. The proteomes of all sequenced 11 predatory bacteria, including two de novo sequenced genomes, and 19 non-predatory bacteria from across the phylogenetic and ecological landscapes were compared. Protein families discriminating between the two groups were identified and quantified, demonstrating that differences in the proteomes of predatory and non-predatory bacteria are large and significant. This analysis allows predictions to be made, as we show by confirming from genome data an over-looked bacterial predator. The predatome exhibits deficiencies in riboflavin and amino acids biosynthesis, suggesting that predators obtain them from their prey. In contrast, these genomes are highly enriched in adhesins, proteases and particular metabolic proteins, used for binding to, processing and consuming prey, respectively. -

Reportable Disease Surveillance in Virginia, 2013
Reportable Disease Surveillance in Virginia, 2013 Marissa J. Levine, MD, MPH State Health Commissioner Report Production Team: Division of Surveillance and Investigation, Division of Disease Prevention, Division of Environmental Epidemiology, and Division of Immunization Virginia Department of Health Post Office Box 2448 Richmond, Virginia 23218 www.vdh.virginia.gov ACKNOWLEDGEMENT In addition to the employees of the work units listed below, the Office of Epidemiology would like to acknowledge the contributions of all those engaged in disease surveillance and control activities across the state throughout the year. We appreciate the commitment to public health of all epidemiology staff in local and district health departments and the Regional and Central Offices, as well as the conscientious work of nurses, environmental health specialists, infection preventionists, physicians, laboratory staff, and administrators. These persons report or manage disease surveillance data on an ongoing basis and diligently strive to control morbidity in Virginia. This report would not be possible without the efforts of all those who collect and follow up on morbidity reports. Divisions in the Virginia Department of Health Office of Epidemiology Disease Prevention Telephone: 804-864-7964 Environmental Epidemiology Telephone: 804-864-8182 Immunization Telephone: 804-864-8055 Surveillance and Investigation Telephone: 804-864-8141 TABLE OF CONTENTS INTRODUCTION Introduction ......................................................................................................................................1 -

There Ae Currently Three Fully Sequenced Deltaproteobacteria
עבודת גמר )תזה( לתואר Thesis for the degree דוקטור לפילוסופיה Doctor of Philosophy מוגשת למועצה המדעית של Submitted to the Scientific Council of the מכון ויצמן למדע Weizmann Institute of Science רחובות, ישראל Rehovot, Israel מאת By אופיר אבידן Ofir Avidan התגובה הראשונית של Bdellovibrio לטרף Early response of Bdellovibrio to its prey מנחים: :Advisors פרופסור שמואל פיטרוקובסקי )מכון Professor Shmuel Pietrokovski וויצמן( (WIS) פרופסור אדוארד יורקביץ Professor Edouard Jurkevitch )האוניברסיטה העברית בירושלים( The Hebrew University of) Jerusalem) חודש ושנה עבריים Month and Year שבט, תשע"ו January, 2016 Table of contents 1. List of abbreviations .................................................................................... 2 2. Abstract................................................................................................................ 3 4 ...................................................................................................................... תקציר .3 4. Introduction ...................................................................................................... 5 5. Research aims ................................................................................................ 12 6. Materials and methods ............................................................................ 13 7. Results ............................................................................................................... 18 7.1 Genetic manipulations in Bdellovibrio ................................................................ -

New 16S Rrna Primers to Uncover Bdellovibrio and Like Organisms Diversity and Abundance Jade Ezzedine, Cécile Chardon, Stéphan Jacquet
New 16S rRNA primers to uncover Bdellovibrio and like organisms diversity and abundance Jade Ezzedine, Cécile Chardon, Stéphan Jacquet To cite this version: Jade Ezzedine, Cécile Chardon, Stéphan Jacquet. New 16S rRNA primers to uncover Bdellovibrio and like organisms diversity and abundance. Journal of Microbiological Methods, Elsevier, 2020, 10.1016/j.mimet.2020.105996. hal-02935301 HAL Id: hal-02935301 https://hal.inrae.fr/hal-02935301 Submitted on 10 Sep 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Journal of Microbiological Methods 175 (2020) 105996 Contents lists available at ScienceDirect Journal of Microbiological Methods journal homepage: www.elsevier.com/locate/jmicmeth New 16S rRNA primers to uncover Bdellovibrio and like organisms diversity T and abundance ⁎ Jade A. Ezzedine, Cécile Chardon, Stéphan Jacquet Université Savoie Mont-Blanc, INRAE, UMR CARRTEL, Thonon-les-Bains, France ARTICLE INFO ABSTRACT Keywords: Appropriate use and specific primers are important in assessing the diversity and abundance of microbial groups Bdellovibrio and like organisms of interest. Bdellovibrio and like organisms (BALOs), that refer to obligate Gram-negative bacterial predators of Primer design other Gram-negative bacteria, evolved in terms of taxonomy and classification over the past two decades. -
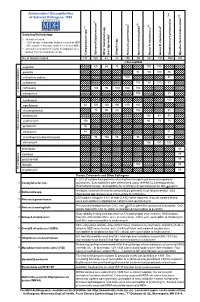
Antimicrobial Susceptibilities of Selected Pathogens, 1999
✔ Antimicrobial Susceptibilities * * † 7 of Selected Pathogens, 1999 8 e † a 2 ✔ ✔ † 3 ✔ † 4 5 6 culosis * r 1 urium spp. m L spp. Sampling Methodology 2 L † all isolates tested * ~ 20% sample of statewide isolates received at MDH spp. ~10% sample of statewide isolates received at MDH Salmonella ** all isolates tested from 7-county metropolitan area oup A streptococci ✔ oup B streptococci r isolates from a normally sterile site r Other (non-typhoidal) G Campylobacter Salmonella typhi Shigella Neisseria gonorrhoeae Neisseria meningitidis G Streptococcus pneumoni Mycobacterium tube No. of Isolates Tested 131 160 43 20 250 55 162 192 559 163 123456 123456% Susceptib123456le 123456123456 123456 123451623456 123451623456 ampicillin 1234561234566012345686 123456 15123456123456 100 100 123456123456 123451623451623451623451623456 123456 penicillin 123456123456123456123456123456 98100 100 76 123456 123456123456123456123456123456123456123456123456 123456 123451623451623451623456 123451623451623456 123456 cefuroxime sodium 123456123456123456123456 100123456123456123456 81 123456 123456123456123456123456123456123456123456123456 123456 cefotaxime 123451623451623451623451623456 100 100100 83 123456 123456123456123456123456123456 123456123456123456123456 123456 123456123456123456123456 ceftriaxone 123456 100 95 100 100 100 123451623451623451623456 -lactam antibiotics 123456123456123456123456123456 123456123456123456123456 β 123451623451623451623451623456 123451623456 123456 meropenem 123456123456123456123456123456 100 123456123456 83 123456 123456123456123456123456123456123456123456123456 -

Microbial NAD Metabolism: Lessons from Comparative Genomics
Dartmouth College Dartmouth Digital Commons Dartmouth Scholarship Faculty Work 9-2009 Microbial NAD Metabolism: Lessons from Comparative Genomics Francesca Gazzaniga Rebecca Stebbins Sheila Z. Chang Mark A. McPeek Dartmouth College Charles Brenner Carver College of Medicine Follow this and additional works at: https://digitalcommons.dartmouth.edu/facoa Part of the Biochemistry Commons, Genetics and Genomics Commons, Medicine and Health Sciences Commons, and the Microbiology Commons Dartmouth Digital Commons Citation Gazzaniga, Francesca; Stebbins, Rebecca; Chang, Sheila Z.; McPeek, Mark A.; and Brenner, Charles, "Microbial NAD Metabolism: Lessons from Comparative Genomics" (2009). Dartmouth Scholarship. 1191. https://digitalcommons.dartmouth.edu/facoa/1191 This Article is brought to you for free and open access by the Faculty Work at Dartmouth Digital Commons. It has been accepted for inclusion in Dartmouth Scholarship by an authorized administrator of Dartmouth Digital Commons. For more information, please contact [email protected]. MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS, Sept. 2009, p. 529–541 Vol. 73, No. 3 1092-2172/09/$08.00ϩ0 doi:10.1128/MMBR.00042-08 Copyright © 2009, American Society for Microbiology. All Rights Reserved. Microbial NAD Metabolism: Lessons from Comparative Genomics Francesca Gazzaniga,1,2 Rebecca Stebbins,1,2 Sheila Z. Chang,1,2 Mark A. McPeek,2 and Charles Brenner1,3* Departments of Genetics and Biochemistry and Norris Cotton Cancer Center, Dartmouth Medical School, Lebanon, New Hampshire -

Impact of Sideways and Bottom-Up Control Factors on Bacterial Community Succession Over a Tidal Cycle
Impact of sideways and bottom-up control factors on bacterial community succession over a tidal cycle Ashvini Chauhan1, Jennifer Cherrier, and Henry N. Williams Environmental Sciences Institute, Florida A&M University, Frederick S. Humphries Science Research Building, Suite 305-D, Tallahassee, FL 32307 Edited by David M. Karl, University of Hawaii, Honolulu, HI, and approved January 22, 2009 (received for review October 21, 2008) In aquatic systems, bacterial community succession is a function of tandem, top-down factors shape bacterial community structure top-down and bottom-up factors, but little information exists on through a variety of processes, including protistan grazing (10) and ‘‘sideways’’ controls, such as bacterial predation by Bdellovibrio- viral lysis (11). Therefore, in all likelihood at any given time, like organisms (BLOs), which likely impacts nutrient cycling within combinations of these factors drive bacterial community succession the microbial loop and eventual export to higher trophic groups. in aquatic ecosystems (12–14). Here we report transient response of estuarine microbiota and BLO Recently, Mou et al. (2) proposed that in marine systems, spp. to tidal-associated dissolved organic matter supply in a river- transient changes in the dissolved organic carbon (DOC) pool are dominated estuary, Apalachicola Bay, Florida. Both dissolved or- less critical in structuring bacterial communities than those that ganic carbon and dissolved organic nitrogen concentrations oscil- result from viral lysis, protistan grazing, or even physicochemical lated over the course of the tidal cycle with relatively higher conditions. Both grazing and viral lyses are selective, such that concentrations observed at low tide. Concurrent with the shift in factors including nonsusceptibility, morphology, size, and motility dissolved organic matter (DOM) supply at low tide, a synchronous offer protection to certain bacterial groups (12). -

The Old Testament Is Dying a Diagnosis and Recommended Treatment 1St Edition Download Free
THE OLD TESTAMENT IS DYING A DIAGNOSIS AND RECOMMENDED TREATMENT 1ST EDITION DOWNLOAD FREE Brent A Strawn | 9780801048883 | | | | | David T. Lamb Strawn offers a few other concrete suggestions about how to save the Old Testament, illustrating several of these by looking at the book of Deuteronomy as a model for second language acquisition. Retrieved 27 August The United States' Centers for Disease Control and Prevention CDC currently recommend that individuals who have been diagnosed and treated for gonorrhea avoid sexual contact with others until at least one week past the final day of treatment in order to prevent the spread of the bacterium. Brent Strawn reminds us of the Old Testament's important role in Christian faith and practice, criticizes current misunderstandings that contribute to its neglect, and offers ways to revitalize its use in the church. None, burning with urinationvaginal dischargedischarge from the penispelvic paintesticular pain [1]. Stunted language learners either: leave faith behind altogether; remain Christian, but look to other resources for how to live their lives; or balkanize in communities that prefer to speak something akin to baby talk — a pidgin-like form of the Old Testament and Bible as a whole — or, worse still, some sort of creole. Geoff, thanks for the reference. Log in. The guest easily identified the passage The Old Testament Is Dying A Diagnosis and Recommended Treatment 1st edition the New Testament, but the Old Testament passage was a swing, and a miss. Instead, our system considers things like how recent a review is and if the reviewer bought the item on Amazon. -

Cultivation of Bdellovibrios
Special Instructions Cultivation of Bdellovibrios Bdellovibrios are unique microorganisms that prey upon a wide variety of susceptible Gram-negative bacteria. Their predatory life style is characterized by two distinct phases, a free-living attack phase and an intraperiplasmic growth phase. Although members of the order Bdellovibrionales are phenotypically quite similar, they do not form a coherent phylogenetic group. Currently, they are classified into five different genera, Bdellovibrio, Bacteriovorax, Peredibacter, Halobacteriovorax and Pseudobacteriovorax. Bdellovibrios can be found in a wide variety of habitats, ranging from soil to sewage, provided these environments are densely populated with bacteria. Some strains represent prey-independent mutants that have lost their requirement for prey cells and hence are facultatively predacious (e.g., Bacteriovorax stolpii DSM 12778) or adapted to an obligate saprophytic life-style (e.g., Bdellovibrio bacteriovorus DSM 12732). Prey-dependent bdellovibrios are usually sensitive to lyophilization and consequently delivered as actively growing cultures from the DSMZ. Fresh samples of attack phase bdellovibrios are shipped either on double-layered agar plates or in liquid broth culture. Presumably, due to the high endogenous respiration rates of bdellovibrios, their viability rapidly decreases after complete lysis of prey cells. Therefore, it is important to transfer the obtained cultures immediately upon receipt into freshly prepared media containing suspensions of susceptible prey cells. An axenic culture of prey cells is shipped along with prey-dependent strains of bdellovibrios. A detailed description of the cultivation of Bdellovibrio bacteriovorus DSM 50701T follows below to exemplify the recommended handling of prey-dependent strains. You will receive from the DSMZ a double layer agar plate of DSMZ medium 257 containing predator and prey cells in the top layer and a tube of slant agar (DSMZ medium 54) with an axenic culture of the prey bacterium Pseudomonas sp. -

1 Supplementary Material a Major Clade of Prokaryotes with Ancient
Supplementary Material A major clade of prokaryotes with ancient adaptations to life on land Fabia U. Battistuzzi and S. Blair Hedges Data assembly and phylogenetic analyses Protein data set: Amino acid sequences of 25 protein-coding genes (“proteins”) were concatenated in an alignment of 18,586 amino acid sites and 283 species. These proteins included: 15 ribosomal proteins (RPL1, 2, 3, 5, 6, 11, 13, 16; RPS2, 3, 4, 5, 7, 9, 11), four genes (RNA polymerase alpha, beta, and gamma subunits, Transcription antitermination factor NusG) from the functional category of Transcription, three proteins (Elongation factor G, Elongation factor Tu, Translation initiation factor IF2) of the Translation, Ribosomal Structure and Biogenesis functional category, one protein (DNA polymerase III, beta subunit) of the DNA Replication, Recombination and repair category, one protein (Preprotein translocase SecY) of the Cell Motility and Secretion category, and one protein (O-sialoglycoprotein endopeptidase) of the Posttranslational Modification, Protein Turnover, Chaperones category, as annotated in the Cluster of Orthologous Groups (COG) (Tatusov et al. 2001). After removal of multiple strains of the same species, GBlocks 0.91b (Castresana 2000) was applied to each protein in the concatenation to delete poorly aligned sites (i.e., sites with gaps in more than 50% of the species and conserved in less than 50% of the species) with the following parameters: minimum number of sequences for a conserved position: 110, minimum number of sequences for a flank position: 110, maximum number of contiguous non-conserved positions: 32000, allowed gap positions: with half. The signal-to-noise ratio was determined by altering the “minimum length of a block” parameter. -

Bdellovibrio and Like Organisms in Lake Geneva: an Unseen Elephant in the Room?
fmicb-11-00098 February 12, 2020 Time: 17:55 # 1 ORIGINAL RESEARCH published: 14 February 2020 doi: 10.3389/fmicb.2020.00098 Bdellovibrio and Like Organisms in Lake Geneva: An Unseen Elephant in the Room? Jade A. Ezzedine1, Louis Jacas1, Yves Desdevises2 and Stéphan Jacquet1* 1 Université Savoie Mont-Blanc, INRAE, CARRTEL, Thonon-les-Bains, France, 2 CNRS, Biologie Intégrative des Organismes Marins, Observatoire Océanologique, Sorbonne Université, Banyuls-sur-Mer, France When considering microbial biotic interactions, viruses as well as eukaryotic grazers are known to be important components of aquatic microbial food webs. It might be the same for bacterivorous bacteria but these groups have been comparatively less studied. This is typically the case of the Bdellovibrio and like organisms (BALOs), which are obligate bacterial predators of other bacteria. Recently, the abundance and distribution Edited by: of three families of this functional group were investigated in perialpine lakes, revealing Susan Fearn Koval, their presence and quantitative importance. Here, a more in-depth analysis is provided University of Western Ontario, Canada for Lake Geneva regarding the diversity of these bacterial predators at different seasons, Reviewed by: Martin W. Hahn, sites and depths. We reveal a seasonal and spatial (vertical) pattern for BALOs. They University of Innsbruck, Austria were also found to be relatively diverse (especially Bdellovibrionaceae) and assigned to Hans-Peter Grossart, both known and unknown phylogenetic clusters. At last we found that most BALOs Leibniz-Institute of Freshwater Ecology and Inland Fisheries (IGB), were positively correlated to other bacterial groups, mainly Gram-negative, in particular Germany Myxococcales (among which many are predators of other microbes).