Metabolic Processes of Methanococcus Maripaludis and Potential Applications Nishu Goyal1, Zhi Zhou2* and Iftekhar A
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Diversity of Understudied Archaeal and Bacterial Populations of Yellowstone National Park: from Genes to Genomes Daniel Colman
University of New Mexico UNM Digital Repository Biology ETDs Electronic Theses and Dissertations 7-1-2015 Diversity of understudied archaeal and bacterial populations of Yellowstone National Park: from genes to genomes Daniel Colman Follow this and additional works at: https://digitalrepository.unm.edu/biol_etds Recommended Citation Colman, Daniel. "Diversity of understudied archaeal and bacterial populations of Yellowstone National Park: from genes to genomes." (2015). https://digitalrepository.unm.edu/biol_etds/18 This Dissertation is brought to you for free and open access by the Electronic Theses and Dissertations at UNM Digital Repository. It has been accepted for inclusion in Biology ETDs by an authorized administrator of UNM Digital Repository. For more information, please contact [email protected]. Daniel Robert Colman Candidate Biology Department This dissertation is approved, and it is acceptable in quality and form for publication: Approved by the Dissertation Committee: Cristina Takacs-Vesbach , Chairperson Robert Sinsabaugh Laura Crossey Diana Northup i Diversity of understudied archaeal and bacterial populations from Yellowstone National Park: from genes to genomes by Daniel Robert Colman B.S. Biology, University of New Mexico, 2009 DISSERTATION Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Biology The University of New Mexico Albuquerque, New Mexico July 2015 ii DEDICATION I would like to dedicate this dissertation to my late grandfather, Kenneth Leo Colman, associate professor of Animal Science in the Wool laboratory at Montana State University, who even very near the end of his earthly tenure, thought it pertinent to quiz my knowledge of oxidized nitrogen compounds. He was a man of great curiosity about the natural world, and to whom I owe an acknowledgement for his legacy of intellectual (and actual) wanderlust. -

Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline
Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline • Phlogenetics of Archaea • Phlogenetics of archaeal lipids • Papers Phyla • Two? main phyla – Euryarchaeota • Methanogens • Extreme halophiles • Extreme thermophiles • Sulfate-reducing – Crenarchaeota • Extreme thermophiles – Korarchaeota? • Hyperthermophiles • indicated only by environmental DNA sequences – Nanoarchaeum? • N. equitans a fast evolving euryarchaeal lineage, not novel, early diverging archaeal phylum – Ancient archael group? • In deepest brances of Crenarchaea? Euryarchaea? Archaeal Lipids • Methanogens – Di- and tetra-ethers of glycerol and isoprenoid alcohols – Core mostly archaeol or caldarchaeol – Core sometimes sn-2- or Images removed due to sn-3-hydroxyarchaeol or copyright considerations. macrocyclic archaeol –PMI • Halophiles – Similar to methanogens – Exclusively synthesize bacterioruberin • Marine Crenarchaea Depositional Archaeal Lipids Biological Origin Environment Crocetane methanotrophs? methane seeps? methanogens, PMI (2,6,10,15,19-pentamethylicosane) methanotrophs hypersaline, anoxic Squalane hypersaline? C31-C40 head-to-head isoprenoids Smit & Mushegian • “Lost” enzymes of MVA pathway must exist – Phosphomevalonate kinase (PMK) – Diphosphomevalonate decarboxylase – Isopentenyl diphosphate isomerase (IPPI) Kaneda et al. 2001 Rohdich et al. 2001 Boucher et al. • Isoprenoid biosynthesis of archaea evolved through a combination of processes – Co-option of ancestral enzymes – Modification of enzymatic specificity – Orthologous and non-orthologous gene -

Geomicrobiological Processes in Extreme Environments: a Review
202 Articles by Hailiang Dong1, 2 and Bingsong Yu1,3 Geomicrobiological processes in extreme environments: A review 1 Geomicrobiology Laboratory, China University of Geosciences, Beijing, 100083, China. 2 Department of Geology, Miami University, Oxford, OH, 45056, USA. Email: [email protected] 3 School of Earth Sciences, China University of Geosciences, Beijing, 100083, China. The last decade has seen an extraordinary growth of and Mancinelli, 2001). These unique conditions have selected Geomicrobiology. Microorganisms have been studied in unique microorganisms and novel metabolic functions. Readers are directed to recent review papers (Kieft and Phelps, 1997; Pedersen, numerous extreme environments on Earth, ranging from 1997; Krumholz, 2000; Pedersen, 2000; Rothschild and crystalline rocks from the deep subsurface, ancient Mancinelli, 2001; Amend and Teske, 2005; Fredrickson and Balk- sedimentary rocks and hypersaline lakes, to dry deserts will, 2006). A recent study suggests the importance of pressure in the origination of life and biomolecules (Sharma et al., 2002). In and deep-ocean hydrothermal vent systems. In light of this short review and in light of some most recent developments, this recent progress, we review several currently active we focus on two specific aspects: novel metabolic functions and research frontiers: deep continental subsurface micro- energy sources. biology, microbial ecology in saline lakes, microbial Some metabolic functions of continental subsurface formation of dolomite, geomicrobiology in dry deserts, microorganisms fossil DNA and its use in recovery of paleoenviron- Because of the unique geochemical, hydrological, and geological mental conditions, and geomicrobiology of oceans. conditions of the deep subsurface, microorganisms from these envi- Throughout this article we emphasize geomicrobiological ronments are different from surface organisms in their metabolic processes in these extreme environments. -

Archaeology of Eukaryotic DNA Replication
Downloaded from http://cshperspectives.cshlp.org/ on September 25, 2021 - Published by Cold Spring Harbor Laboratory Press Archaeology of Eukaryotic DNA Replication Kira S. Makarova and Eugene V. Koonin National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, Maryland 20894 Correspondence: [email protected] Recent advances in the characterization of the archaeal DNA replication system together with comparative genomic analysis have led to the identification of several previously un- characterized archaeal proteins involved in replication and currently reveal a nearly com- plete correspondence between the components of the archaeal and eukaryotic replication machineries. It can be inferred that the archaeal ancestor of eukaryotes and even the last common ancestor of all extant archaea possessed replication machineries that were compa- rable in complexity to the eukaryotic replication system. The eukaryotic replication system encompasses multiple paralogs of ancestral components such that heteromeric complexes in eukaryotes replace archaeal homomeric complexes, apparently along with subfunctionali- zation of the eukaryotic complex subunits. In the archaea, parallel, lineage-specific dupli- cations of many genes encoding replication machinery components are detectable as well; most of these archaeal paralogs remain to be functionally characterized. The archaeal rep- lication system shows remarkable plasticity whereby even some essential components such as DNA polymerase and single-stranded DNA-binding protein are displaced by unrelated proteins with analogous activities in some lineages. ouble-stranded DNA is the molecule that Okazaki fragments (Kornberg and Baker 2005; Dcarries genetic information in all cellular Barry and Bell 2006; Hamdan and Richardson life-forms; thus, replication of this genetic ma- 2009; Hamdan and van Oijen 2010). -

Insights Into Archaeal Evolution and Symbiosis from the Genomes of a Nanoarchaeon and Its Inferred Crenarchaeal Host from Obsidian Pool, Yellowstone National Park
University of Tennessee, Knoxville TRACE: Tennessee Research and Creative Exchange Microbiology Publications and Other Works Microbiology 4-22-2013 Insights into archaeal evolution and symbiosis from the genomes of a nanoarchaeon and its inferred crenarchaeal host from Obsidian Pool, Yellowstone National Park Mircea Podar University of Tennessee - Knoxville, [email protected] Kira S. Makarova National Institutes of Health David E. Graham University of Tennessee - Knoxville, [email protected] Yuri I. Wolf National Institutes of Health Eugene V. Koonin National Institutes of Health See next page for additional authors Follow this and additional works at: https://trace.tennessee.edu/utk_micrpubs Part of the Microbiology Commons Recommended Citation Biology Direct 2013, 8:9 doi:10.1186/1745-6150-8-9 This Article is brought to you for free and open access by the Microbiology at TRACE: Tennessee Research and Creative Exchange. It has been accepted for inclusion in Microbiology Publications and Other Works by an authorized administrator of TRACE: Tennessee Research and Creative Exchange. For more information, please contact [email protected]. Authors Mircea Podar, Kira S. Makarova, David E. Graham, Yuri I. Wolf, Eugene V. Koonin, and Anna-Louise Reysenbach This article is available at TRACE: Tennessee Research and Creative Exchange: https://trace.tennessee.edu/ utk_micrpubs/44 Podar et al. Biology Direct 2013, 8:9 http://www.biology-direct.com/content/8/1/9 RESEARCH Open Access Insights into archaeal evolution and symbiosis from the genomes of a nanoarchaeon and its inferred crenarchaeal host from Obsidian Pool, Yellowstone National Park Mircea Podar1,2*, Kira S Makarova3, David E Graham1,2, Yuri I Wolf3, Eugene V Koonin3 and Anna-Louise Reysenbach4 Abstract Background: A single cultured marine organism, Nanoarchaeum equitans, represents the Nanoarchaeota branch of symbiotic Archaea, with a highly reduced genome and unusual features such as multiple split genes. -
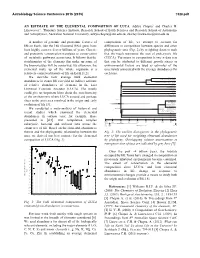
An Estimate of the Elemental Composition of Luca
Astrobiology Science Conference 2015 (2015) 7328.pdf AN ESTIMATE OF THE ELEMENTAL COMPOSITION OF LUCA. Aditya Chopra1 and Charles H. Lineweaver1, 1Planetary Science Institute, Research School of Earth Sciences and Research School of Astronomy and Astrophysics, Australian National University, [email protected], [email protected] A number of genomic and proteomic features of composition of life, we attempt to account for life on Earth, like the 16S ribosomal RNA gene, have differences in composition between species and other been highly conserved over billions of years. Genetic phylogenetic taxa (Fig. 2) by weighting datasets such and proteomic conservation translates to conservation that the result represents the root of prokaryotic life of metabolic pathways across taxa. It follows that the (LUCA). Variations in composition between data sets stoichiometry of the elements that make up some of that can be attributed to different growth stages or the biomolecules will be conserved. By extension, the environmental factors are used as estimates of the elemental make up of the whole organism is a uncertainty associated with the average abundances for relatively conserved feature of life on Earth [1,2]. each taxa. We describe how average bulk elemental Euryarchaeota 1a Methanococci, Methanobacteria, Methanopyri 7 Euryarchaeota 1b abundances in extant life can yield an indirect estimate 6 Thermoplasmata, Methanomicrobia, Halobacteria, Archaeoglobi Euryarchaeota 2 4 Thermococci of relative abundances of elements in the Last Crenarchaeota Sulfolobus, Thermoproteus ? Thaumarchaeota Universal Common Ancestor (LUCA). The results Cenarchaeum ? Korarchaeota ARMAN could give us important hints about the stoichiometry ? 2 Archaeal Richmond Mine Acidophilic Nanoorganisms Nanoarchaeota of the environment where LUCA existed and perhaps Archaea Terrabacteria clues to the processes involved in the origin and early Actinobacteria, Deinococcus-Thermus, 1 Cyanobacteria, Life Chloroflexi, 9 evolution of life [3]. -

Brian W. Waters
INVESTIGATION OF 2-OXOACID OXIDOREDUCTASES IN METHANOCOCCUS MARIPALUDIS AND LARGE-SCALE GROWTH OF M. MARIPALUDIS by BRIAN W. WATERS (Under the direction of Dr. William B. Whitman) ABSTRACT With the emergence of Methanococcus maripaludis as a genetic model for methanogens, it becomes imperative to devise inexpensive yet effective ways to grow large numbers of cells for protein studies. Chapter 2 details methods that were used to cut costs of growing M. maripaludis in large scale. Also, large scale growth under different conditions was explored in order to find the conditions that yield the most cells. Chapter 3 details the phylogenetic analysis of 2-oxoacid oxidoreductase (OR) homologs from M. maripaludis. ORs are enzymes that catalyze the oxidative decarboxylation of 2-oxoacids to their acyl-CoA derivatives in many prokaryotes. Also in chapter 3, a specific OR homolog in M. maripaludis, an indolepyruvate oxidoreductase (IOR), was mutagenized, and the mutant was characterized. PCR and Southern hybridization analysis showed gene replacement. A no-growth phenotype on media containing aromatic amino acid derivatives was found. INDEX WORDS: Methanococcus maripaludis, fermentor, 2-oxoacid oxidoreductase, indolepyruvate oxidoreductase INVESTIGATION OF 2-OXOACID OXIDOREDUCTASES IN METHANOCOCCUS MARIPALUDIS AND LARGE-SCALE GROWTH OF M. MARIPALUDIS by BRIAN W. WATERS B.S., The University of Georgia,1999 A Thesis Submitted to the Graduate Faculty of The University of Georgia in Partial Fulfillment of the Requirements for the Degree MASTER OF SCIENCE ATHENS, GEORGIA 2002 © 2002 Brian W. Waters All Rights Reserved INVESTIGATION OF 2-OXOACID OXIDOREDUCTASES IN METHANOCOCCUS MARIPALUDIS AND LARGE-SCALE GROWTH OF M. MARIPALUDIS by BRIAN W. -

Fermentation and Anaerobic Decomposition in a Hot Spring
Fermentation and anaerobic decomposition in a hot spring microbial mat by Karen Leigh Anderson A thesis submitted in partial fulfillment of requirements for the degree of Master of Science in Microbiology Montana State University © Copyright by Karen Leigh Anderson (1984) Abstract: Fermentation was investigated in a low sulfate hot spring microbial mat (Octopus Spring) according to current models on anaerobic decomposition. The mat was studied to determine what fermentation products accumulated, where in the mat they accumulated, and what factors affected their accumulation. Mat samples were incubated under dark anaerobic conditions to measure accumulation of fermentation products. Acetate and propionate (ca. 3:1) were the major products to accumulate in a 55°,C mat. Other products accumulated to a much lesser extent. Incubation of mat samples of varying thickness showed that fermentation occurred in the top 4mm of the mat. This has interesting implications for fermentative organisms in the mat due to the diurnal changes in mat oxygen concentrations. Fermentation measured in mat samples collected at various temperatures (50°,-70°C) showed acetate and propionate to be the major accumulation products. According to the interspecies hydrogen transfer model, the hydrogen concentration in a system affects the types of fermentation products produced. At a 65° C site, with natural high hydrogen levels, and at a 55°C site, with active methanogenesis, fermentation product accumulation was compared. There was a greater ratio of reduced fermentation products to acetate, with the exception of propionate, at 65°C. Ethanol accumulated at the 65°C site, as did lactate, though to a lesser extent. -

Exploring the Nutritional Values of Hydrogenotrophic Bacteria As Space Food
ISSN: 2455-2631 © December 2018 IJSDR | Volume 3, Issue 12 Exploring the Nutritional Values of Hydrogenotrophic Bacteria as Space Food 1M.GIRISH, 2S.ABIRAMI, 3Dr.V.MAHALAKSHMI 1,2Research scholar, 3Professor 1,3Madras Christian College, 2V.R.R Institute of Biomedical Sciences. Abstract: Hydrogenotrophs are organisms that are capable of metabolizing molecular hydrogen as source of energy and hence hydrogenotrophy is performed by carbon dioxide reducing organism to produce methane. Methanogens are diverse group of strict anaerobes where methanogenesis is a major route in the anaerobic mineralization of organic matter. They have very unique enzyme and cofactors involved in this metabolic pathway. The anaerobic chamber provides a convenient culture system for large-scale studies of strictly anaerobic and/or facultative anaerobic bacteria. In this study, the Hydrogenotrophic organism isolated was characterized as Methanosarcina sp. Though these hydrogenotrophs utilise carbon dioxide and grow in a minimal growth requirement, they were found to be rich in carbohydrates and protein content. Therefore, Hydrogenotroph being a good food source, can be used for future space travel where the astronauts could carry them aboard and synthesis their own food. The by-products produced by these organisms can be used to produce a protein rich meal. This system can be incorporated on to space craft where this works as a closed loop between astronauts and hydrogenotrophs, where carbon dioxide released by astronauts can be captured and filtered through a system and supplied for the growth of these microbes which in turn synthesis carbohydrates. Keywords: Hydrogenotrophs, space food, anaerobes. I. INTRODUCTION Hydrogenotrophs are organisms that are capable of metabolizing molecular hydrogen as source of energy. -

Tackling the Methanopyrus Kandleri Paradox Céline Brochier*, Patrick Forterre† and Simonetta Gribaldo†
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by PubMed Central Open Access Research2004BrochieretVolume al. 5, Issue 3, Article R17 Archaeal phylogeny based on proteins of the transcription and comment translation machineries: tackling the Methanopyrus kandleri paradox Céline Brochier*, Patrick Forterre† and Simonetta Gribaldo† Addresses: *Equipe Phylogénomique, Université Aix-Marseille I, Centre Saint-Charles, 13331 Marseille Cedex 3, France. †Institut de Génétique et Microbiologie, CNRS UMR 8621, Université Paris-Sud, 91405 Orsay, France. reviews Correspondence: Céline Brochier. E-mail: [email protected] Published: 26 February 2004 Received: 14 November 2003 Revised: 5 January 2004 Genome Biology 2004, 5:R17 Accepted: 21 January 2004 The electronic version of this article is the complete one and can be found online at http://genomebiology.com/2004/5/3/R17 reports © 2004 Brochier et al.; licensee BioMed Central Ltd. This is an Open Access article: verbatim copying and redistribution of this article are permitted in all media for any purpose, provided this notice is preserved along with the article's original URL. ArchaealPhylogeneticsequencedusingrespectively). two phylogeny concatenated genomes, analysis based it of is datasetsthe now on Archaea proteinspossible consisting has ofto been thetest of transcription alternative mainly14 proteins established approach involv and translationed byes in 16S bytranscription rRNAusing machineries: largesequence andsequence 53comparison.tackling ribosomal datasets. the Methanopyrus Withproteins We theanalyzed accumulation(3,275 archaealkandleri and 6,377 of phyparadox comp positions,logenyletely Abstract deposited research Background: Phylogenetic analysis of the Archaea has been mainly established by 16S rRNA sequence comparison. With the accumulation of completely sequenced genomes, it is now possible to test alternative approaches by using large sequence datasets. -

WARREN LYNWOOD GARDNER Expression Vectors for the Methane-Producing Archaeon Methanococcus Maripaludis (Under the Direction of WILLIAM B
WARREN LYNWOOD GARDNER Expression vectors for the methane-producing archaeon Methanococcus maripaludis (Under the Direction of WILLIAM B. WHITMAN) Methanogens are strict anaerobes that use one and two carbon compounds for methanogenesis. They contain many unusual cofactors and enzymes, and many of their proteins are oxygen-sensitive and are present at low concentrations within the cells. An expression system that overexpresses homologous and heterologous proteins would facilitate research on the unique oxygen-sensitive enzymes in these organisms. However, traditional expression systems may lack the cofactors and maturation enzymes necessary for the expression of the active holoenzymes. Therefore, a major goal of this work was to develop expression vectors. To develop a shuttle vector, a series of integrative vectors were first constructed. The integrative vectors were based on the pUC-derivative pMEB.2. pMEB.2 provided the puromycin resistance marker for methanococci, an Escherichia coli origin of replication, and an ampicillin resistance marker for E. coli. A multiple cloning site and the Methanococcus voltae histone promoter (PhmvA) were added to form the integrative, expression vector pWLG14. To demonstrate the utility of PhmvA, pWLG14 was used to overexpress the genomic copy of the Methanococcus maripaludis acetohydroxyacid synthase. To form an expression shuttle vector suitable for heterologous genes, pWLG14 and the cryptic plasmid pURB500 from M. maripaludis strain were ligated together to form pWLG30. pWLG30 was the first expression shuttle vector for the methanogens. To demonstrate the utility of pWLG30, the E. coli b-galactosidase gene was cloned into pWLG30 to yield pWLG30+lacZ. Upon transformation into M. maripaludis, the recombinant strain expressed b-galactosidase to the level of 1% of the cellular protein. -

Conversion of Amazon Rainforest to Agriculture Alters Community Traits of Methane-Cycling Organisms
Conversion of Amazon rainforest to agriculture alters community traits of methane-cycling organisms Kyle M. Meyer1***, Ann M. Klein1**, Jorge L.M. Rodrigues2*, Klaus Nüsslein3, Susannah Tringe4, Babur Mirza5, James M. Tiedje6, Titus Brown7 and Brendan J.M. Bohannan1* 1. Institute of Ecology and Evolution, Department of Biology, University of Oregon, Eugene, OR 2. Department of Land, Air and Water Resources, University of California, Davis, CA 3. Department of Microbiology, University of Massachusetts, Amherst, MA 4. Joint Genome Institute, United States Department of Energy, Walnut Creek, CA 5. 6. Department of Plant, Soil, and Microbial Sciences, Michigan State University, East Lansing, MI 7. Department of Population Health and Reproduction, University of California, Davis, CA * Corresponding authors ([email protected], [email protected], [email protected]) **These authors contributed equally to this work. ABSTRACT Land use change is one of the greatest environmental threats worldwide, especially to tropical forests. The Amazon rainforest has been subject to particularly high rates of land use change, primarily to cattle pasture. A commonly observed response to cattle pasture establishment in the Amazon is the conversion of soil from a methane sink in rainforest, to a methane source in pasture. However, it is not known how the microorganisms that mediate methane flux are altered by land use change. Here we use the deepest metagenomic sequencing of Amazonian soil to date to investigate differences in methane-cycling microorganisms and their traits across rainforest and cattle pasture soils. We found that methane-cycling microorganisms responded to land use change, with the strongest responses exhibited by methane- consuming, rather than methane-producing, microorganisms.