Assessment of Mahseer Population Structure and Genetic Variation Using Atpase6
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Forest of Madhya Pradesh
Build Your Own Success Story! FOREST OF MADHYA PRADESH As per the report (ISFR) MP has the largest forest cover in the country followed by Arunachal Pradesh and Chhattisgarh. Forest Cover (Area-wise): Madhya Pradesh> Arunachal Pradesh> Chhattisgarh> Odisha> Maharashtra. Forest Cover (Percentage): Mizoram (85.4%)> Arunachal Pradesh (79.63%)> Meghalaya (76.33%) According to India State of Forest Report the recorded forest area of the state is 94,689 sq. km which is 30.72% of its geographical area. According to Indian state of forest Report (ISFR – 2019) the total forest cover in M.P. increased to 77,482.49 sq km which is 25.14% of the states geographical area. The forest area in MP is increased by 68.49 sq km. The first forest policy of Madhya Pradesh was made in 1952 and the second forest policy was made in 2005. Madhya Pradesh has a total of 925 forest villages of which 98 forest villages are deserted or located in national part and sanctuaries. MP is the first state to nationalise 100% of the forests. Among the districts, Balaghat has the densest forest cover, with 53.44 per cent of its area covered by forests. Ujjain (0.59 per cent) has the least forest cover among the districts In terms of forest canopy density classes: Very dense forest covers an area of 6676 sq km (2.17%) of the geograhical area. Moderately dense forest covers an area of 34, 341 sqkm (11.14% of geograhical area). Open forest covers an area of 36, 465 sq km (11.83% of geographical area) Madhya Pradesh has 0.06 sq km. -

Master of Philosophy in GEOLOGY
Studies on the Seasonal Fluctuation of Water Levels and Seasonal Changes in Chemical Quality of Ground Water in Kota District Rajasthan. DISSERTATION SUBMITTED IN PARTIAL FULFILMENT OF THE REQUIREMENTS FOR THE AWARD OF THE DECREE OF Master of Philosophy IN GEOLOGY BY SHAMiM AHMAD Under the Supervision of Dr; Shadab Khurshid DEPARTMENT OF GEOLOGY ALIGARH MUSLIM UNIVERSITY ALIGARH (INDIA) 1987 DS1082 D£PART.v£NT OF GEOLOGY ALIGAHK M.,> _i .: , \i\ERSiTY ALiG\K:i l\:y.ni DR. SHADAB KHURSHID 16- 6- S)^ M.ScPG Dip. Hydrogeo.,M.Phil.,Ph.D. ^^'' TO WHOM IT MAY CONCERN This is to certify that the dissertation entitled, "Studies on the Seasonal Fluctuation of Water Levels and Seasonal Changes in Cherracal Quality of Ground Water in Kota district,Rajasthan", is the original contribution of Mr. Shamim Ahmad in the field of Hydrogeology which was carried out under my supervision. It has not been published in part or full anywhere else. Mr. Shamim Ahmad is allowed to submit: this work for the award of M.Phil, degree of Aiigarh Muslim University, Aiigarh. ( SHADAB KHURSHID 1 SUPERVISOR :ONTENTS LIST OF TABLES 1 LIST OF FIGURES iv CHAPTER-I INTRODUCTION 1 General Statement 1 Aim and Scope 3 Methods of Study 3 Hydrogeological Data 4 Fauna and Flora 4 Location and Communication 5 CHAPTER-II GEOLOGY OF THE AREA 7 Physiography 7 Drainage Pattern 7 Stratigraphy 8 Vindhyan Supergroup 10 CHAPTER-III LAND USB PATTERN AND CROPS 14 Characteristics and Properties of Soil 16 in Canal Comman area CHAPTER-IV GROUND WATER LE\i^:L AND FLUCTUATION 25 Ground Water Characteristics -

Dewas District Madhya Pradesh
DEWAS DISTRICT MADHYA PRADESH Ministry of Water Resources Central Ground Water Board North Central Region BHOPAL 2013 DISTRICT PROFILE - DEWAS DISTRICT S.No. ITEMS Statistics 1. GENERAL INFORMATION i) Geographical area (sq. km) 7020.84 ii) Administrative Divisions (As on 2011 ) Number of Tehsil 6 Number of Blocks 6 Number of Panchayats 497 Number of Villages 1127 iii)Population (Census 2011) 289438 iv)Normal Rainfall (mm) 1083 2. GEOMORPHOLOGY i) Major Physiographic Units 1.Dewas Plateau 2. Kali Sindh Basin 3. Vindhyan Range 4. Middle Narmada Valley Kshipra sub-basin Kali Sindh sub-basin ii) Major Drainage Chotti Kali Sindh sub-basin Kanhar sub-basin Khari sub-basin Datuni sub-basin Jamner sub-basin Narmada direct catchment 3. LAND USE (ha) i) Forest area: 206600 ii) Net area sown: 388400 iii) Cultivable area: 624500 4. MAJOR SOIL TYPES Black cotton , Sandy loam, Clayey loam,, Murram 5. AREA UNDER PRINCIPAL CROPS (2009) Wheat, Soyabean, Groundnut, Cotton, etc 6. IRRIGATION BY DIFFERENT SOURCES No of Area (ha) Structures Dug wells 36531 65900 Tube wells/Bore wells 23119 93100 Tanks/Ponds 169 4760 Canals 15 6760 Other Sources - 11044 Net Irrigated Area - 193640 7. NUMBER OF GROUND WATER MONITORING WELLS OF CGWB (As on 31.3.2013) No. of Dug Wells 16 No. of Piezometers 11 8 PREDOMINANT GEOLOGICAL Deccan trap lava flows FORMATIONS 9 HYDROGEOLOGY Major Water Bearing Formation Weathered/Fractured (Pre-monsoon depth to water level during Basalt 2012) 2.90 – 24.47 mbgl (Post-monsoon depth to water level during 0.06 – 15.19 mbgl 2012) Long Term water level trend in 10 years (2003- 0.007 to 2.74 m (Rise) 2012) in m/yr 0.109 to 0.27 m (Fall) 10. -

Annuity Project
Public Private Partnership in Annuity Project DRAFT CONCESSION AGREEMENT VOLUME-II: SCHEDULES FOR RSHIP Package-1 : Development and Maintenance of Kanwas -Khanpur - Aklera section of SH-74, Deoli- Kanwas section of SH-74 A, Alot (MP) – Gangdhar- Suwansara (MP) section of SH-19 C and Kherli-Pahari section of SH-22 (Total length: 175.36 Km) under Design, Build, Operate/ Maintain and Transfer on Annuity Mode. Public Works Department Government of Rajasthan SCHEDULE-A (See Clause 10.1) SITE OF THE PROJECT 1 The Site 1.1 Site of the Intermediate-Lane/ Two Lane Project shall include the land, buildings, structures and road works as described in Annex-I of this Schedule-A. 1.2 An inventory of the Site including the land, buildings, structures, road works, trees and any other immovable property on, or attached to, the Site shall be prepared jointly by the Authority Representative and the Concessionaire, and such inventory shall form part of the memorandum referred to in Clause 10.3.1 of the Agreement. 1.3 Additional land required for the Project has been described in Annex-I of this Schedule-A. This land shall be acquired by the Authority, at its own cost and upon acquisition, such land shall form part of the Site and vest in the Authority. 2 Annex-I (Schedule-A) Site for Project Highway-1: Kanwas -Khanpur -Aklera section of SH-74 1. SITE The Site of the Intermediate Lane Highway-1 comprises the section of State Highway No.74 as under: Kanwas-Khanpur-Aklera section: Commencing from km 11.730 at Awan Junction (with SH 74 A), Near Kanwas to km 86.950 at Junction with NH-52, near Arnia in the State of Rajasthan having existing total length of 75.22 kms The land, carriageway and structures comprising the Site are described below. -

LIFELINES of INDIA's CIVILISATION in India, a River Is a Mini-Cosmos In
LIFELINES OF INDIA’S CIVILISATION In India, a river is a mini-cosmos in concept. Every river is a mother deity who spawns mythology, art, dance, music, architecture, history and spirituality. Each one has a clear identity, appearance, value, style and spirit just like a beautiful woman. In every age, diverse human communities have reinvented themselves on river-banks with fascinating nuances…. ‘Her shimmering gold-and-white garments dazzle like a thousand suns. The jewels in her crown shine like the crescent moon. Her smiling face lights up the whole world. In her hands, she carries a pot of nectar, a symbol of immortality. Her lotus-fresh presence brings a sense of purity and joy to all beings….’. At first glance, this reads like an over-the-top flowery description of a beautiful woman coined by some besotted lover. But to those conversant with the fascinating river-lore of India, this is the mythical portrayal of the River Ganga, written by Sage Valmiki, author of India’s immortal epic Ramayan. It describes the celestial Ganga as she descends from the heavens to the earth to bring salvation to mankind. This story, known asGangavataran, is such a fundamental tenet of Indian culture that it has held countless generations of Indians in awe for millenniums. The Ganga, arguably the most picturised and written-about river in the world, has been called the Mother of India’s Spirituality and has been immortalized in sculpture, art, literature, poetry, music and dance. Following her descent to the mortal world to sanctify human efforts to attain salvation, the Ganga is perceived as mokshdayini, the Mother Goddess whose waters bring relief from sin, sorrow and suffering. -

Assessment of Domestic Pollution Load from Urban Agglomeration in Ganga Basin: Madhya Pradesh
Report Code: 063_GBP_IIT_EQP_S&R_13_VER 1_DEC 2014 Assessment of Domestic Pollution Load from Urban Agglomeration in Ganga Basin: Madhya Pradesh GRBMP: Ganga River Basin Management Plan by Indian Institutes of Technology IIT IIT IIT IIT IIT IIT IIT Bombay Delhi Guwahati Kanpur Kharagpur Madras Roorkee Report Code: 063_GBP_IIT_EQP_S&R_13_VER 1_DEC 2014 2 Report Code: 063_GBP_IIT_EQP_S&R_13_VER 1_DEC 2014 Preface In exercise of the powers conferred by sub-sections (1) and (3) of Section 3 of the Environment (Protection) Act, 1986 (29 of 1986), the Central Government has constituted National Ganga River Basin Authority (NGRBA) as a planning, financing, monitoring and coordinating authority for strengthening the collective efforts of the Central and State Government for effective abatement of pollution and conservation of the river Ganga. One of the important functions of the NGRBA is to prepare and implement a Ganga River Basin Management Plan (GRBMP). A Consortium of 7 Indian Institute of Technology (IIT) has been given the responsibility of preparing Ganga River Basin Management Plan (GRBMP) by the Ministry of Environment and Forests (MoEF), GOI, New Delhi. Memorandum of Agreement (MoA) has been signed between 7 IITs (Bombay, Delhi, Guwahati, Kanpur, Kharagpur, Madras and Roorkee) and MoEF for this purpose on July 6, 2010. This report is one of the many reports prepared by IITs to describe the strategy, information, methodology, analysis and suggestions and recommendations in developing Ganga River Basin Management Plan (GRBMP). The overall Frame Work for documentation of GRBMP and Indexing of Reports is presented on the inside cover page. There are two aspects to the development of GRBMP. -

Madhya Pradesh: Geography Contents
MPPSCADDA Web: mppscadda.com Telegram: t.me/mppscadda WhatsApp/Call: 9953733830, 7982862964 MADHYA PRADESH: GEOGRAPHY CONTENTS ❖ Chapter 1 Introduction to Geography of Madhya Pradesh ❖ Chapter 2 Physiographic Divisions of Madhya Pradesh ❖ Chapter 3 Climate Season and Rainfall in Madhya Pradesh ❖ Chapter 4 Soils of Madhya Pradesh ❖ Chapter 5 Rivers and Drainage System of Madhya Pradesh ❖ Chapter 6 Major Irrigation and Electrical Projects of Madhya Pradesh ❖ Chapter 7 Forests and Forest Produce of Madhya Pradesh ❖ Chapter 8 Biodiversity of Madhya Pradesh CONTACT US AT: Website :mppscadda.com Telegram :t.me/mppscadda WhatsApp :7982862964 WhatsApp/Call :9711733833 Gmail: [email protected] FREE TESTS: http://mppscadda.com/login/ Web: mppscadda.com Telegram: t.me/mppscadda WhatsApp/Call: 9953733830, 7982862964 INTRODUCTION TO GEOGRAPHY OF MADHYA PRADESH MPPSCADDA Web: mppscadda.com Telegram: t.me/mppscadda WhatsApp/Call: 9953733830, 7982862964 1. INTRODUCTION TO GEOGRAPHY OF MADHYA PRADESH Topography of Madhya Pradesh • Madhya Pradesh is situated at the north-central part of Peninsular plateau India, whose boundary can be classified in the north by the plains of Ganga-Yamuna, in the west by the Aravalli, east by the Chhattisgarh plain and in the south by the Tapti Valley and the plateau of Maharashtra. • Geological Structure: Geologically MP is a part of Gondwana Land. 3,08,252 km2 Area (9.38% of the total area of India) 21⁰ 6' - 26 ⁰30' Latitudinal Expansion 605 km (North to South) 74⁰ 59' - 82 ⁰66' Longitudinal Expansion 870 km (East to West) Width is more than Length Indian Standard Meridian Singrauli District ( Only one district in MP) 82⁰30' passes • Topic of Cancer and Indian Standard Meridian do not cross each other in any part of MP Geographical Position of MP • Madhya Pradesh is the 2nd (second) largest state by area with its area 9.38% of the total area of the country. -
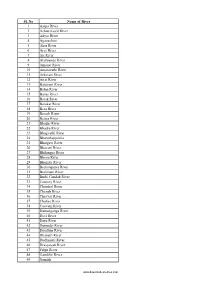
List of Rivers in India
Sl. No Name of River 1 Aarpa River 2 Achan Kovil River 3 Adyar River 4 Aganashini 5 Ahar River 6 Ajay River 7 Aji River 8 Alaknanda River 9 Amanat River 10 Amaravathi River 11 Arkavati River 12 Atrai River 13 Baitarani River 14 Balan River 15 Banas River 16 Barak River 17 Barakar River 18 Beas River 19 Berach River 20 Betwa River 21 Bhadar River 22 Bhadra River 23 Bhagirathi River 24 Bharathappuzha 25 Bhargavi River 26 Bhavani River 27 Bhilangna River 28 Bhima River 29 Bhugdoi River 30 Brahmaputra River 31 Brahmani River 32 Burhi Gandak River 33 Cauvery River 34 Chambal River 35 Chenab River 36 Cheyyar River 37 Chaliya River 38 Coovum River 39 Damanganga River 40 Devi River 41 Daya River 42 Damodar River 43 Doodhna River 44 Dhansiri River 45 Dudhimati River 46 Dravyavati River 47 Falgu River 48 Gambhir River 49 Gandak www.downloadexcelfiles.com 50 Ganges River 51 Ganges River 52 Gayathripuzha 53 Ghaggar River 54 Ghaghara River 55 Ghataprabha 56 Girija River 57 Girna River 58 Godavari River 59 Gomti River 60 Gunjavni River 61 Halali River 62 Hoogli River 63 Hindon River 64 gursuti river 65 IB River 66 Indus River 67 Indravati River 68 Indrayani River 69 Jaldhaka 70 Jhelum River 71 Jayamangali River 72 Jambhira River 73 Kabini River 74 Kadalundi River 75 Kaagini River 76 Kali River- Gujarat 77 Kali River- Karnataka 78 Kali River- Uttarakhand 79 Kali River- Uttar Pradesh 80 Kali Sindh River 81 Kaliasote River 82 Karmanasha 83 Karban River 84 Kallada River 85 Kallayi River 86 Kalpathipuzha 87 Kameng River 88 Kanhan River 89 Kamla River 90 -

Rivers of India
Downloaded From examtrix.com Compilation of Rivers www.onlyias.in Mahanadi RiverDownloaded From examtrix.com Source: Danadkarnya Left bank: Sheonath, Hasdo and Mand Right bank: Tel, Jonk, Ong Hirakund dam Olive Ridley Turtles: Gahirmatha beach, Orissa: Nesting turtles River flows through the states of Chhattisgarh and Odisha. River Ends in Bay of Bengal Mahanadi RiverDownloaded From examtrix.com Mahanadi RiverDownloaded From examtrix.com • The Mahanadi basin extends over states of Chhattisgarh and Odisha and comparatively smaller portions of Jharkhand, Maharashtra and Madhya Pradesh, draining an area of 1.4 lakh Sq.km. • It is bounded by the Central India hills on the north, by the Eastern Ghats on the south and east and by the Maikala range on the west. • The Mahanadi (“Great River”) follows a total course of 560 miles (900 km). • It has its source in the northern foothills of Dandakaranya in Raipur District of Chhattisgarh at an elevation of 442 m. • The Mahanadi is one of the major rivers of the peninsular rivers, in water potential and flood producing capacity, it ranks second to the Godavari. Mahanadi RiverDownloaded From examtrix.com • Other small streams between the Mahanadi and the Rushikulya draining directly into the Chilka Lake also forms the part of the basin. • After receiving the Seonath River, it turns east and enters Odisha state. • At Sambalpur the Hirakud Dam (one of the largest dams in India) on the river has formed a man-made lake 35 miles (55 km) long. • It enters the Odisha plains near Cuttack and enters the Bay of Bengal at False Point by several channels. -

Replenishment Study Report for River Bed Mining Project of Minor Mineral Ordinary Sand (Bajri)
REPLENISHMENT STUDY REPORT FOR RIVER BED MINING PROJECT OF MINOR MINERAL ORDINARY SAND (BAJRI) AT RIVERS, NALLAS, GAIR MUMKIN NADI IN TEHSIL – GANGDHAR, DISTRICT – JHALAWAR, STATE – RAJASTHAN NAME OF RIVER – CHAMBAL , SIPRA, CHOTI KALI SINDH, CHACBARAM RIVER & THEIR CONNECTED FLOW NALLAHS Proposed production – 7,50,000 TPA LEASE AREA – 1179.84 Hect. CATEGORY- ΄A΄ EC letter No. – J-11015/327/2013-IA.II (M) dt. 26/02/2016 Replenishment Study Period – Pre – monsoon and Post – monsoon season of year 2017 APPLICANT PREPARED BY INDRAJEET SINGH JHALA M/S UDAIPUR MIN-TECH PVT. LTD. NALA MOHALLA, KHARI KUIYA, 206-APEKSHA COMPLEX, SECTOR NO.-11, P.O. JHALAWAR, TEHSIL – JHALRAPATAN HIRAN MAGARI, UDAIPUR-313002 (RAJ.). DIST. – JHALAWAR (RAJ.) PH- 91-294-2489672 (OFF.) Mob. +91 9414193514 (M) Mob. 9414167672 E mail:- [email protected] E mail :- [email protected] ACCREDITED BY NABET (S.NO. 144) CATEGORY “A” FOR SECTOR 1 1 CONTENTS SR. NO. PARTICULARS PAGE NO. 1. INTRODUCTION 3 2. PROJECT DETAIL 4 3. LOCATION MAP 6 4. KEY PLAN 7 5. CATCHMENT AREA MAP 8 6. PHOTOGRAPHS OF APPLIED LEASE AREA 9 7. STAGES OF ENVIRONMENTAL CLEARANCE 10 8. REPLENISHMENT STUDY 11 2 Introduction: The proposed project is for River Bed mining of ordinary sand (Bajri) from River Chambal, Sipra, Choti Kali Sindh, Chacbaram River & their connected flow nallahs. The applied lease area is situated in Tehsil – Gangdhar of District – Jhalawar State – Rajasthan. Applicant: Mr. Indrajeet Singh Jhala, Nala Mohalla, Khari Kuiya, P.O. Jhalawar, Tehsil – Jhalarapatan, District – Jhalawar, (Rajasthan) Mob. +91 9414193514 Mining of Ordinary Sand (Bajri) is proposed in the river bed, the lease area is 1179.84 ha (Govt. -

Incredible Results in IAS 2013 5 Ranks 62 Ranks in Top 50 Ranks in the Final List
9 1 0 2 - Y A M Justice Bobde panel gives clean chit to CJI in sexual harassment probe A case for Judicial Self-examination Image source : Indian Legal Intellectual Property Control of Transboundary South-South MNC’s control vs Movements of co-operation over data- Ways, Farmer Rights Hazardous Wastes Means and Dominance RESULTS Incredible results in IAS 2013 5 Ranks 62 Ranks in Top 50 Ranks in the final list Rank 9 Rank 12 Rank 23 Rank 40 Rank 46 Divyanshu Jha Neha Jain Prabhav joshi Gaurang Rathi Udita Singh We broke our past record in IAS 2014 6 Ranks 12 Ranks 83 Ranks in Top 50 in Top 100 Overall Selections Rank 4 Rank 5 Rank 16 Rank 23 Rank 28 Rank 39 Vandana Rao Suharsha Bhagat Ananya Das Anil Dhameliya Kushaal Yadav Vivekanand T.S We did it again in IAS 2015 5 Ranks 14 Ranks 162 Ranks in Top 50 in Top 100 In The Final List Rank 20 Rank 24 Rank 25 Rank 27 Rank 47 Vipin Garg Khumanthem Chandra Pulkit Garg Anshul Diana Devi Mohan Garg Agarwal And we’ve done it yet again in IAS 2016 8 Ranks 18 Ranks 215 Ranks in Top 50 in Top 100 In The Final List Rank 2 Rank 5 Rank 12 Rank 30 Rank 32 Anmol Sher Abhilash Tejaswi Prabhash Avdhesh Singh Bedi Mishra Rana Kumar Meena And we’ve done it yet again in IAS 2017 5 Ranks 34 Ranks 236 Ranks in Top 10 in Top 100 In The Final List Rank 3 Rank 6 Rank 8 Rank 9 Rank 10 Sachin Koya sree Anubhav Saumya Abhishek Gupta Harsha Singh Sharma Surana Ashima Abhijeet Varjeet Keerthi Utsav Gaurav Abhilash Vikramaditya Vishal Mittal Sinha Walia Vasan V Gautam Kumar Baranwal Singh Malik Mishra Rank-12 Rank-19 Rank-21 -

1 | Page BRIEF SUMMARY of the PROJECT 1. Introduction of The
Brief Summary BRIEF SUMMARY OF THE PROJECT 1. Introduction of the project Shri Chandra Pal Singh S/o Shri Ganga Singh has proposed for River Bed Mining Project of Bajri, Production Capacity 1.31 MTPA from Gair Mumkin Nadi & Nallah Area-252.48 ha, Tehsil-Digod & Pipalda, District: Kota & Tehsil Anta, District: Baran (Rajasthan). Letter of Intent (LOI) for grant of mining lease for Bajri over an area of 252.48 ha has been issued by the Superintending Mining Engineer, Directorate of Mines & Geology, Government of Rajasthan vide letter no. Directorate/P2 (A.3)/Kota/Bajri/2012/3007 dated 24.12.2013. 2. Screening Category As per EIA Notification dated 14 th Sep, 2006 amended as on date, the project falls under S. No ‘1’ (Mining of Minerals), Project or Activity ‘1(a) - (3)’, Category “A”. 3. Brief Description of Project a. Location Villages: Haripura, Bijavta, Naravta, Khedli, Nonera, Kolana, Pipalda Kalan, Dhibri, Bankiya, Barana, Bislai, Dhansuri, Bhunen, Kherula, Khedli Parasram, Khedli Tawran, Bagawda, Khyavda, Barod, Patonda, Raipura, Baldada, Palaytha, Bijora. Tehsil: Pipalda, Digod & Anta District: Kota & Baran State: Rajasthan 4. Environmental Setting S. No. Particulars Details 1 Nearest National Highway/ NH -76 is crossing the lease area in East to West State Highway direction SH-70 is crossing to the lease area in East to West direction 2 Inter District Boundary Bundi & Kota ( crossing parallel to the lease area ) 3 Nearest Railway Station Anta ( ~ 5.5 km in East direction) 4 Nearest Airport Kota ( ~38 km in West direction) 5 National