Technical, Microbial, and Economic Study on Thermophilic Solid-State Anaerobic
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

METABOLIC EVOLUTION in GALDIERIA SULPHURARIA By
METABOLIC EVOLUTION IN GALDIERIA SULPHURARIA By CHAD M. TERNES Bachelor of Science in Botany Oklahoma State University Stillwater, Oklahoma 2009 Submitted to the Faculty of the Graduate College of the Oklahoma State University in partial fulfillment of the requirements for the Degree of DOCTOR OF PHILOSOPHY May, 2015 METABOLIC EVOLUTION IN GALDIERIA SUPHURARIA Dissertation Approved: Dr. Gerald Schoenknecht Dissertation Adviser Dr. David Meinke Dr. Andrew Doust Dr. Patricia Canaan ii Name: CHAD M. TERNES Date of Degree: MAY, 2015 Title of Study: METABOLIC EVOLUTION IN GALDIERIA SULPHURARIA Major Field: PLANT SCIENCE Abstract: The thermoacidophilic, unicellular, red alga Galdieria sulphuraria possesses characteristics, including salt and heavy metal tolerance, unsurpassed by any other alga. Like most plastid bearing eukaryotes, G. sulphuraria can grow photoautotrophically. Additionally, it can also grow solely as a heterotroph, which results in the cessation of photosynthetic pigment biosynthesis. The ability to grow heterotrophically is likely correlated with G. sulphuraria ’s broad capacity for carbon metabolism, which rivals that of fungi. Annotation of the metabolic pathways encoded by the genome of G. sulphuraria revealed several pathways that are uncharacteristic for plants and algae, even red algae. Phylogenetic analyses of the enzymes underlying the metabolic pathways suggest multiple instances of horizontal gene transfer, in addition to endosymbiotic gene transfer and conservation through ancestry. Although some metabolic pathways as a whole appear to be retained through ancestry, genes encoding individual enzymes within a pathway were substituted by genes that were acquired horizontally from other domains of life. Thus, metabolic pathways in G. sulphuraria appear to be composed of a ‘metabolic patchwork’, underscored by a mosaic of genes resulting from multiple evolutionary processes. -

Metagenomic Shotgun Sequencing Analysis of Canalicular Concretions in Lacrimal Canaliculitis Cases
Article Metagenomic Shotgun Sequencing Analysis of Canalicular Concretions in Lacrimal Canaliculitis Cases Yukinobu Okajima 1,*, Takashi Suzuki 1, Chika Miyazaki 2, Satoshi Goto 3, Sho Ishikawa 4 , Yuka Suzuki 1, Kotaro Aoki 5 , Yoshikazu Ishii 5, Kazuhiro Tateda 5 and Yuichi Hori 1 1 Department of Ophthalmology, Toho University, 6-11-1 Omori-nishi, Ota-ku, Tokyo 143-8541, Japan; [email protected] (T.S.); [email protected] (Y.S.); [email protected] (Y.H.) 2 Hyogo Prefectural Amagasaki General Medical Center, 2-17-77 Higashi-nanba cho, Amagasaki 661-0892, Japan; [email protected] 3 Department of Ophthalmology, School of Medicine, The Jikei University, 3-19-18 Shinbashi-nishi, Minato-ku, Tokyo 105-8471, Japan; [email protected] 4 Department of Ophthalmology, School of Medicine, Saitama University, 38 Morohongo Moroyama-machi, Iruma-gun, Saitama 350-0495, Japan; [email protected] 5 Department of Microbiology and Infectious Diseases, School of Medicine, Toho University, 6-11-1 Omori-nishi, Ota-ku, Tokyo 143-8541, Japan; [email protected] (K.A.); [email protected] (Y.I.); [email protected] (K.T.) * Correspondence: [email protected]; Tel.: +81-3-3762-4151; Fax: +81-3-3298-0030 Abstract: Lacrimal canaliculitis is a rare infection of the lacrimal canaliculi with canalicular con- cretions formed by aggregation of organisms. Metagenomic shotgun sequencing analysis using next-generation sequencing has been used to detect pathogens directly from clinical samples. Using Citation: Okajima, Y.; Suzuki, T.; this technology, we report cases of successful pathogen detection of canalicular concretions in lacrimal Miyazaki, C.; Goto, S.; Ishikawa, S.; canaliculitis cases. -

Interplay of Microbial Communities with Mineral Environments in Coralline Algae
Science of the Total Environment 757 (2021) 143877 Contents lists available at ScienceDirect Science of the Total Environment journal homepage: www.elsevier.com/locate/scitotenv Interplay of microbial communities with mineral environments in coralline algae Patricia M. Valdespino-Castillo a,1, Andrea Bautista-García b,1, Fabio Favoretto b,c, Martín Merino-Ibarra d, Rocío J. Alcántara-Hernández e, Teresa Pi-Puig e,f, F. Sergio Castillo d, Silvia Espinosa-Matías g, Hoi-Ying Holman a, Anidia Blanco-Jarvio b,⁎ a Molecular Biophysics and Integrated Bioimaging Division, Lawrence Berkeley National Laboratory, Berkeley, CA, United States b Laboratorio de Bioingeniería y Ciencias Ambientales (BICA), Departamento Académico de Ingeniería en Pesquerías, Universidad Autónoma de Baja California Sur, La Paz, BCS, Mexico c Gulf of California Marine Program, Scripps Institution of Oceanography, University of California San Diego, CA, United States d Unidad Académica de Biodiversidad Acuática, Instituto de Ciencias del Mar y Limnología, Universidad Nacional Autónoma de México, Mexico City, Mexico e Instituto de Geología, Universidad Nacional Autónoma de México, Mexico City, Mexico f Laboratorio Nacional de Geoquímica y Mineralogía (LANGEM), Universidad Nacional Autónoma de México, Mexico City, Mexico g Laboratorio de Microscopía Electrónica de Barrido, Facultad de Ciencias, Universidad Nacional Autónoma de México, Mexico City, Mexico HIGHLIGHTS GRAPHICAL ABSTRACT • The interplay of microorganisms and algal mineral bioconstructions remains poorly understood. • Carbonates rich in Fe and Mg found make CA relevant targets to study coastal resilience. • Halophiles and evaporite minerals con- currently suggest halophilic microenvi- ronments in the thallus. • Bacterial microbiota correlated signifi- cantly with temperature and nutrients. • Key bacteria might play relevant roles in adaptive responses of coralline algae. -
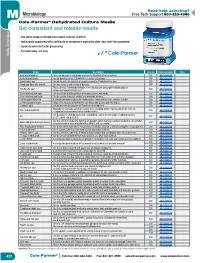
Get Consistent and Reliable Results
Need help selecting? M Microbiology Free Tech Support 800-323-4340 Cole-Parmer® Dehydrated Culture Media Get consistent and reliable results – Use when ready or dehydrated culture extends shelf life – Individually numbered with certificate of compliance expiration date, and shelf-life guarantee – Quick dissolve for faster processing – For laboratory use only Media, Dehydrated Media, Media Description Size (g) Catalog number Price Agar, bacteriological This clear gel agar is high grade and manufactured using the ice method 500 GH-14200-10 Azide dextrose broth Use for detection of fecal Streptococci in water and sewage 500 GH-14202-00 Baird parker agar Use for the selective isolation of coagulase positive Staphylococci in food 500 GH-14202-01 Blood agar base with low pH Use to cultivate a wide variety of bacteria 500 GH-14202-02 This is used as a differential medium in the isolation and presumptive identification of Bile Esculin agar 500 GH-14202-03 enterococci/group D streptococci Brain heart infusion agar Use to cultivate a wide spectrum of bacteria, yeasts, and molds 500 GH-14200-12 Brain heart infusion broth Use to cultivate fastidious aerobic and anaerobic bacteria 500 GH-14202-05 Brilliant green bile broth A standard methods medium for confirmed and completed tests for coliforms in water 500 GH-14200-14 Buffered peptone water Helps in the recovery of Salmonella from foods after preservation techniques 500 GH-14202-08 Cetrimide agar Use for the selective isolation of Pseudomonas aeruginosa 500 GH-14200-52 This broth is especially suited for environmental sampling where neutralization of the chemical D/E neutralizing Broth 500 GH-14202-11 is important to determining the bactericidal activity Use to isolate E. -

Prepared Culture Media
PREPARED CULTURE MEDIA 121517SS PREPARED CULTURE MEDIA Made in the USA AnaeroGRO™ DuoPak A 02 Bovine Blood Agar, 5%, with Esculin 13 AnaeroGRO™ DuoPak B 02 Bovine Blood Agar, 5%, with Esculin/ AnaeroGRO™ BBE Agar 03 MacConkey Biplate 13 AnaeroGRO™ BBE/PEA 03 Bovine Selective Strep Agar 13 AnaeroGRO™ Brucella Agar 03 Brucella Agar with 5% Sheep Blood, Hemin, AnaeroGRO™ Campylobacter and Vitamin K 13 Selective Agar 03 Brucella Broth with 15% Glycerol 13 AnaeroGRO™ CCFA 03 Brucella with H and K/LKV Biplate 14 AnaeroGRO™ Egg Yolk Agar, Modified 03 Buffered Peptone Water 14 AnaeroGRO™ LKV Agar 03 Buffered Peptone Water with 1% AnaeroGRO™ PEA 03 Tween® 20 14 AnaeroGRO™ MultiPak A 04 Buffered NaCl Peptone EP, USP 14 AnaeroGRO™ MultiPak B 04 Butterfield’s Phosphate Buffer 14 AnaeroGRO™ Chopped Meat Broth 05 Campy Cefex Agar, Modified 14 AnaeroGRO™ Chopped Meat Campy CVA Agar 14 Carbohydrate Broth 05 Campy FDA Agar 14 AnaeroGRO™ Chopped Meat Campy, Blood Free, Karmali Agar 14 Glucose Broth 05 Cetrimide Select Agar, USP 14 AnaeroGRO™ Thioglycollate with Hemin and CET/MAC/VJ Triplate 14 Vitamin K (H and K), without Indicator 05 CGB Agar for Cryptococcus 14 Anaerobic PEA 08 Chocolate Agar 15 Baird-Parker Agar 08 Chocolate/Martin Lewis with Barney Miller Medium 08 Lincomycin Biplate 15 BBE Agar 08 CompactDry™ SL 16 BBE Agar/PEA Agar 08 CompactDry™ LS 16 BBE/LKV Biplate 09 CompactDry™ TC 17 BCSA 09 CompactDry™ EC 17 BCYE Agar 09 CompactDry™ YMR 17 BCYE Selective Agar with CAV 09 CompactDry™ ETB 17 BCYE Selective Agar with CCVC 09 CompactDry™ YM 17 BCYE -

Phylogeny of Nitrogenase Structural and Assembly Components Reveals New Insights Into the Origin and Distribution of Nitrogen Fixation Across Bacteria and Archaea
microorganisms Article Phylogeny of Nitrogenase Structural and Assembly Components Reveals New Insights into the Origin and Distribution of Nitrogen Fixation across Bacteria and Archaea Amrit Koirala 1 and Volker S. Brözel 1,2,* 1 Department of Biology and Microbiology, South Dakota State University, Brookings, SD 57006, USA; [email protected] 2 Department of Biochemistry, Genetics and Microbiology, University of Pretoria, Pretoria 0004, South Africa * Correspondence: [email protected]; Tel.: +1-605-688-6144 Abstract: The phylogeny of nitrogenase has only been analyzed using the structural proteins NifHDK. As nifHDKENB has been established as the minimum number of genes necessary for in silico predic- tion of diazotrophy, we present an updated phylogeny of diazotrophs using both structural (NifHDK) and cofactor assembly proteins (NifENB). Annotated Nif sequences were obtained from InterPro from 963 culture-derived genomes. Nif sequences were aligned individually and concatenated to form one NifHDKENB sequence. Phylogenies obtained using PhyML, FastTree, RapidNJ, and ASTRAL from individuals and concatenated protein sequences were compared and analyzed. All six genes were found across the Actinobacteria, Aquificae, Bacteroidetes, Chlorobi, Chloroflexi, Cyanobacteria, Deferribacteres, Firmicutes, Fusobacteria, Nitrospira, Proteobacteria, PVC group, and Spirochaetes, as well as the Euryarchaeota. The phylogenies of individual Nif proteins were very similar to the overall NifHDKENB phylogeny, indicating the assembly proteins have evolved together. Our higher resolution database upheld the three cluster phylogeny, but revealed undocu- Citation: Koirala, A.; Brözel, V.S. mented horizontal gene transfers across phyla. Only 48% of the 325 genera containing all six nif genes Phylogeny of Nitrogenase Structural and Assembly Components Reveals are currently supported by biochemical evidence of diazotrophy. -

View Our Full Water Sampling Vials Product Offering
TABLE OF CONTENTS Environmental Monitoring 1 Sample Prep and Dilution 8 Dehydrated Culture Media - Criterion™ 12 Prepared Culture Media 14 Chromogenic Media - HardyCHROM™ 18 CompactDry™ 20 Quality Control 24 Membrane Filtration 25 Rapid Tests 26 Lab Supplies/Sample Collection 27 Made in the USA Headquarters Distribution Centers 1430 West McCoy Lane Santa Maria, California Santa Maria, CA 93455 Olympia, Washington 800.266.2222 : phone Salt Lake City, Utah [email protected] Phoenix, Arizona HardyDiagnostics.com Dallas, Texas Springboro, Ohio Hardy Diagnostics has a Quality Lake City, Florida Management System that is certified Albany, New York to ISO 13485 and is a FDA licensed Copyright © 2020 Hardy Diagnostics Raleigh, North Carolina medical device manufacturer. Environmental Monitoring Hardy Diagnostics offers a wide selection of quality products intended to help keep you up to date with regulatory compliance, ensure the efficacy of your cleaning protocol, and properly monitor your environment. 800.266.2222 [email protected] HardyDiagnostics.com 1 Environmental Monitoring Air Sampling TRIO.BAS™ Impact Air Samplers introduced the newest generation of microbial air sampling. These ergonomically designed instruments combine precise air sampling with modern connectivity to help you properly assess the air quality of your laboratory and simplify your process. MONO DUO Each kit includes: Each kit includes: TRIO.BAS™ MONO air sampler, induction TRIO.BAS™ DUO air sampler, battery battery charger and cable, aspirating charger -

Microbial Diversity and Cellulosic Capacity in Municipal Waste Sites By
Microbial diversity and cellulosic capacity in municipal waste sites by Rebecca Co A thesis presented to the University of Waterloo in fulfilment of the thesis requirement for the degree of Master of Science in Biology Waterloo, Ontario, Canada, 2019 © Rebecca Co 2019 Author’s Declaration This thesis consists of material all of which I authored or co-authored: see Statement of Contributions included in the thesis. This is a true copy of the thesis, including any required final revisions, as accepted by my examiners. I understand that my thesis may be made electronically available to the public. ii Statement of Contributions In Chapter 2, the sampling and DNA extraction and sequencing of samples (Section 2.2.1 - 2.2.2) were carried out by Dr. Aneisha Collins-Fairclough and Dr. Melessa Ellis. The work described in Section 2.2.3 Metagenomic pipeline and onwards was done by the thesis’s author. Sections 2.2.1 Sample collection – 2.2.4 16S rRNA gene community profile were previously published in Widespread antibiotic, biocide, and metal resistance in microbial communities inhabiting a municipal waste environment and anthropogenically impacted river by Aneisha M. Collins- Fairclough, Rebecca Co, Melessa C. Ellis, and Laura A. Hug. 2018. mSphere: e00346-18. The writing and analyses incorporated into this chapter are by the thesis's author. iii Abstract Cellulose is the most abundant organic compound found on earth. Cellulose’s recalcitrance to hydrolysis is a major limitation to improving the efficiency of industrial applications. The biofuel, pulp and paper, agriculture, and textile industries employ mechanical and chemical methods of breaking down cellulose. -

BD Industry Catalog
PRODUCT CATALOG INDUSTRIAL MICROBIOLOGY BD Diagnostics Diagnostic Systems Table of Contents Table of Contents 1. Dehydrated Culture Media and Ingredients 5. Stains & Reagents 1.1 Dehydrated Culture Media and Ingredients .................................................................3 5.1 Gram Stains (Kits) ......................................................................................................75 1.1.1 Dehydrated Culture Media ......................................................................................... 3 5.2 Stains and Indicators ..................................................................................................75 5 1.1.2 Additives ...................................................................................................................31 5.3. Reagents and Enzymes ..............................................................................................75 1.2 Media and Ingredients ...............................................................................................34 1 6. Identification and Quality Control Products 1.2.1 Enrichments and Enzymes .........................................................................................34 6.1 BBL™ Crystal™ Identification Systems ..........................................................................79 1.2.2 Meat Peptones and Media ........................................................................................35 6.2 BBL™ Dryslide™ ..........................................................................................................80 -

An Evolutionary Network of Genes Present in the Eukaryote Common Ancestor Polls Genomes on Eukaryotic and Mitochondrial Origin
GBE An Evolutionary Network of Genes Present in the Eukaryote Common Ancestor Polls Genomes on Eukaryotic and Mitochondrial Origin Thorsten Thiergart, Giddy Landan, Marc Schenk, Tal Dagan, and William F. Martin* Institute of Molecular Evolution, Heinrich-Heine University Du¨ sseldorf, Germany *Corresponding author: E-mail: [email protected]. Accepted: 14 February 2011 Abstract To test the predictions of competing and mutually exclusive hypotheses for the origin of eukaryotes, we identified from a sample of 27 sequenced eukaryotic and 994 sequenced prokaryotic genomes 571 genes that were present in the eukaryote common ancestor and that have homologues among eubacterial and archaebacterial genomes. Maximum- likelihood trees identified the prokaryotic genomes that most frequently contained genes branching as the sister to the eukaryotic nuclear homologues. Among the archaebacteria, euryarchaeote genomes most frequently harbored the sister to the eukaryotic nuclear gene, whereas among eubacteria, the a-proteobacteria were most frequently represented within the sister group. Only 3 genes out of 571 gave a 3-domain tree. Homologues from a-proteobacterial genomes that branched as the sister to nuclear genes were found more frequently in genomes of facultatively anaerobic members of the rhiozobiales and rhodospirilliales than in obligate intracellular ricketttsial parasites. Following a-proteobacteria, the most frequent eubacterial sister lineages were c-proteobacteria, d-proteobacteria, and firmicutes, which were also the prokaryote genomes least frequently found as monophyletic groups in our trees. Although all 22 higher prokaryotic taxa sampled (crenarchaeotes, c-proteobacteria, spirochaetes, chlamydias, etc.) harbor genes that branch as the sister to homologues present in the eukaryotic common ancestor, that is not evidence of 22 different prokaryotic cells participating at eukaryote origins because prokaryotic ‘‘lineages’’ have laterally acquired genes for more than 1.5 billion years since eukaryote origins. -

150259-ID-None.Pdf
DIPONEGORO JOURNAL OF MAQUARES Volume 5, Nomor 3, Tahun 2016, Halaman: 157-164 MANAGEMENT OF AQUATIC RESOURCES http://ejournal-s1.undip.ac.id/index.php/maquares ANALISIS TOTAL BAKTERI COLIFORM DI PERAIRAN MUARA KALI WISO JEPARA The Analysis of Total Coliform Bacteria in Kali Wiso Estuary Jepara Wiwid Widyaningsih, Supriharyono *), Niniek Widyorini Program Studi Manajemen Sumberdaya Perairan Fakultas Perikanan dan Ilmu Kelautan, Universitas Diponegoro Jl. Prof. Soedarto, SH, Tembalang, Semarang, Jawa Tengah – 50275, Telp/Fax. +6224 7474698 Email : [email protected] ABSTRAK Kali Wiso merupakan sungai yang berada di tengah kota Jepara. Perairan ini menjadi tempat pembuangan limbah-limbah secara langsung. Limbah tersebut diantaranya limbah domestik, limbah pasar, limbah kapal, serta limbah TPI. Berdasarkan masukan limbah tersebut menjadikan muara ini tercemar. Perairan yang tercemar dapat dilihat dari pengamatan secara fisika, kimia, maupun biologis. Kondisi perairan yang tercemar secara biologis dilihat dari keberadaan bakteri patogen yang ada di perairan. Indikator bakteri yang digunakan yaitu bakteri coliform, karena sifatnya yang berkorelasi positif dengan bakteri patogen lainnya. Pemanfaatan perairan ini digunakan untuk kegiatan pelabuhan, tempat bersandar kapal nelayan, serta kegiatan perikanan yang ada di sekitar perairan Jepara. Oleh karena itu perlu diketahui kepadatan bakteri coliform sehingga dapat bermanfaat sesuai dengan peruntukannya. Tujuan dari penelitian ini adalah untuk mengetahui total bakteri coliform serta mengetahui adanya bakteri Escherichia coli. Penelitain ini dilakukan pada bulan Maret 2016 di Muara Kali Wiso dengan dua kali pengulangan dalam kondisi pasang dan surut. Metode yang digunakan yaitu survei dengan teknik sampling purposive sampling. Metode analisa laboratorium yang digunakan berdasarkan SNI -01-2332- 1991. Kepadatan bakteri coliform pada perairan muara Kali Wiso yaitu >110.000 sel/100ml dan bakteri Escherichia coli sebesar >110.000 sel/100ml. -
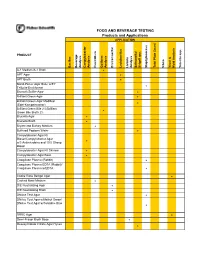
DONE Food and Beverage Testing
FOOD AND BEVERAGE TESTING Products and Applications APPLICATION PRODUCT Bacillus Beverage Analysis Campylobacter Analysis Clostridium Coliform Analysis Environmental Lactobacillus Listeria Analysis Salmonella/ Shigell spp. Staphylococcus Total Plate Count Vibrio Yeast & Mold Analysis Yersinia spp. A-1 Medium/A-1 Broth ! APT Agar ! APT Broth ! Baird-Parker Agar Base w/EY Tellurite Enrichment ! Bismuth Sulfite Agar ! Brilliant Green Agar ! Brilliant Green Agar Modified (Edel-Kampelmacher) ! Brilliant Green Bile 2%/Brilliant Green Bile Broth 2% ! Brucella Agar ! Brucella Broth ! Bryant and Burkey Medium ! Buffered Peptone Water ! Campylobacter Agar Kit Blaser/Campylobacter Agar w/5 Antimicrobics and 10% Sheep ! Blood Campylobacter Agar Kit Skirrow ! Campylobacter Agar Base ! Coagulase Plasma (Rabbit) ! Coagulase Plasma EDTA (Rabbit)/ Coagulase Plasma w/EDTA ! Cooke Rose Bengal Agar ! Cooked Meat Medium ! D/E Neutralizing Agar ! D/E Neutralizing Broth ! DNAse Test Agar ! DNAse Test Agar w/Methyl Green/ DNAse Test Agar w/Toluidine Blue ! DRBC Agar ! Demi-Fraser Broth Base ! Desoxycholate Citrate Agar Hynes ! FOOD AND BEVERAGE TESTING Products and Applications APPLICATION PRODUCT Bacillus Beverage Analysis Campylobacter Analysis Clostridium Coliform Analysis Environmental Lactobacillus Listeria Analysis Salmonella/ Shigell spp. Staphylococcus Total Plate Count Vibrio Yeast & Mold Analysis Yersinia spp. Differential Reinforced Clostridial Agar ! EC Medium/EC Broth ! EC Medium with MUG/EC Broth w/MUG ! Elliker Broth ! m Endo Agar LES ! m Endo