Evaluation of DNA Barcode Libraries Used in the UK and Developing an Action Plan to Fill Priority Gaps
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

CPI Newsletter – June 2018
Mail Stop 2404 • College Station, TX 77843-2404 • (979) 862-9166 • [email protected] ------------------------------------------------------------------------------------ CPI Newsletter – June 2018 ------------------------------------------------------------------------------------ Included in this issue: ⎯ Update from the CPI Chair ⎯ Huffines Faculty Research Seed Grants – Deadline June 15, 2018 ⎯ Potential Opportunity for NSF and NIH Supplements to Existing Grants ⎯ Export Controls Designated Liaison Network ⎯ 3rd Annual Postdoctoral Research Symposium – September 19, 2018 ⎯ Sign Up for Notifications about Limited Submission Proposal Opportunities ⎯ Bulletin for Principal Investigators 2017-18 CPI roster: Chair — Penny Riggs , COALS ● Vice Chair — David Threadgill , Veterinary Medicine ● AgriLife Extension — Craig Carpenter, Gaylon Morgan ● AgriLife Research — Michael Brewer, Ambika Chandra, Fugen Dou, Lee Tarpley ● Architecture — Ergun Akleman ● Bush School — Kent Portney ● COALS — Fuller Bazer, Russell Cross, Martin Dickman, Micky Eubanks, Elizabeth Pierson, David Stelly ● Education – Oi-Man Kwok, Jeffrey Liew ● Engineering — Jorge Alvarado, Ulisses Braga-Neto, Zachary Grasley, Melissa Grunlan, Tony Hsieh Sheng-Jen, Daniel Jiménez, Jodie Lutkenhaus, Ramesh Talreja ● Geosciences — Alejandro Orsi, Pamela Plotkin, Brendan Roark ● IBT/PHARM/RCHI — Julian Hurdle ● Law – Susan Fortney ● Liberal Arts — Sandra Braman, Steve Maren, Harland Prechel ● Mays — Korok Ray ● Medicine —Vytas Bankaitis, Kayla Bayless ● Science — Darren DePoy, Michael Hall, -

Plant Biology '99
THE NEWSLETTER OF THE AMERICAN SOCIETY OF PLANT PHYSIOLOGISTS Volume 26, Number 1 January/February 1999 Plant Biology '99 III Plant lobs Grab Crabs: Chesapeake Boy Crabs Await You 01 Plant Biology '99! Photo courtesy of Baltimore Areo Convention &Visitors Association. Join your colleagues in celebrating ASPP's SYMPOSIA 75th Anniversary at Plant Biology '99 in Baltimore, Maryland, on July 24-28! Auxin Biology Organizers: Mark Estelle and Ottaline Leyser ASPP's 75th Anniversary Meeting Events: Cell Cycle Regulation Organizer: Tom Jacobs Special President's Symposium functional Plant Genomics Banquet Honoring Past Officers, Award Winners, & Organizer: JeffBennetzen ASPP Traditions Biochemical Genetics Organizer: Dean Della Penna Chesapeake Bay Crab & Chicken Feast President's Symposium: Global Issues in Plant Biology ASPP Family Tree Organizer: Brian Larkins Speakers: Peter Raven, Lester Brown, and Extended Poster Sessions Robert Fraley INSIDE ... i T ASPP-Sponsored Workshop in Greece . ' '~. I' T Novartis, UC-Berkeley Plant Scientists Reach $25 Million Research Collaboration T Call for Nominations for Officers and Awards ASPP Future ASPP Annual Meetings OFFICERS & STAFF .CONTENTS flresident Brian A. Larkins .".""""""" """.. 520·621·9958 1 flresident·Elect .·Plant Biology '99 Deborah Delmer """"""" .."""""""""""""""",,, 530·752·7561 1999 Immediate flost flresident ·3 '" . Ken Keegstra .."""""""""""" ..""""""""""""",517·353·2770 Saturday, July 24, through Secretory . ASPP-Sponsored Workshop in Greece Daniel R. Bush ""......,.."""""""""", ..,..""""""", 217·333·6109 Wednesday, July 28 Treasurer ;MbnagingEditor of Plant Physiology Terri Lomax ."""" .."........."""""""""""."""""", 541·737·5278 .Resigns ' Baltimore, Maryland Choir, Boord of Trustees Douglas D. Randall .........".""""""....."..."""""" 573·882·4847 ASPP's 75th anniversary Choir, flublications Committee 4 . Rebella Chasan .."""""""..."......."""""""""""" 202·628·1500 Public Affairs .' '. meeting Choir, Committee on the -NSF's $85 Millibn in Awards Status of Women in fllont flhysio/ogy Ann M. -

Annual Report Fy 2018 Human Frontier Science Program Organization
APRIL 2017 APRIL 2018 — MARCH 2019 ANNUAL REPORT FY 2018 HUMAN FRONTIER SCIENCE PROGRAM ORGANIZATION The Human Frontier Science Program Organization (HFSPO) is unique, supporting international collaboration to undertake innovative, risky, basic research at the frontier of the life sciences. Special emphasis is given to the support and training of independent young investigators, beginning at the postdoctoral level. The Program is implemented by an international organisation, supported financially by Australia, Canada, France, Germany, India, Italy, Japan, the Republic of Korea, New Zealand, Norway, Singapore, Switzerland, the United Kingdom of Great Britain and Nothern Ireland, the United States of America, and the European Commission. Since 1990, over 7000 researchers from more than 70 countries have been supported. Of these, 28 HFSP awardees have gone on to receive the Nobel Prize. 2 The following documents are available on the HFSP website www.hfsp.org: Joint Communiqués (Tokyo 1992, Washington 1997, Berlin 2002, Bern 2004, Ottawa 2007, Canberra 2010, Brussels 2013, London 2016): https://www.hfsp.org/about/governance/membership Statutes of the International Human Frontier Science Program Organization: https://www.hfsp.org/about/governance/hfspo-statutes Guidelines for the participation of new members in HFSPO: https://www.hfsp.org/about/governance/membership General reviews of the HFSP (1996, 2001, 2006-2007, 2010, 2018): https://www.hfsp.org/about/strategy/reviews Updated and previous lists of awards, including titles and abstracts: -

Current Positions
Jay Shendure, MD, PhD Updated December 31, 2020 Current Positions Investigator, Howard Hughes Medical Institute Professor, Genome Sciences, University of Washington Director, Allen Discovery Center for Cell Lineage Director, Brotman Baty Institute for Precision Medicine Contact Information E-mail: [email protected] Lab website: http://krishna.gs.washington.edu Office phone: (206) 685-8543 Education • 2007 M.D., Harvard Medical School (Boston, Massachusetts) • 2005 Ph.D. in Genetics, Harvard University (Cambridge, Massachusetts) Research Advisor: George M. Church Thesis entitled “Multiplex Genome Sequencing and Analysis” • 1996 A.B., summa cum laude in Molecular Biology, Princeton University (Princeton, NJ) Research Advisor: Lee M. Silver Professional Experience • 2017 – present Scientific Director Brotman-Baty Institute for Precision Medicine • 2017 – present Scientific Director Allen Discovery Center for Cell Lineage Tracing • 2015 – present Investigator Howard Hughes Medical Institute • 2015 – present Full Professor (with tenure) Department of Genome Sciences, University of Washington, Seattle, WA • 2010 – present Affiliate Professor Division of Human Biology, Fred Hutchinson Cancer Research Center, Seattle, WA • 2011 – 2015 Associate Professor (with tenure) Department of Genome Sciences, University of Washington, Seattle, WA • 2007 – 2011 Assistant Professor Department of Genome Sciences, University of Washington, Seattle, WA • 1998 – 2007 Medical Scientist Training Program (MSTP) Candidate Department of Genetics, Harvard Medical School, Boston, WA • 1997 – 1998 Research Scientist Vaccine Division, Merck Research Laboratories, Rahway, NJ • 1996 – 1997 Fulbright Scholar to India Department of Pediatrics, Sassoon General Hospital, Pune, India 1 Jay Shendure, MD, PhD Honors, Awards, Named Lectures • 2019 Richard Lounsbery Award (for extraordinary scientific achievement in biology & medicine) National Academy of Sciences • 2019 Jeffrey M. Trent Lectureship in Cancer Research National Human Genome Research Institute, National Institutes of Health • 2019 Paul D. -

NEWSLETTER VOLUME 34, NUMBER 2 Scientists NIH Budget: a Moving Target Adapting to $1.629 Billion: Amount Republicans in the Biomedical Research
ASCB M A R C H 2 0 1 1 NEWSLETTER VOLUME 34, NUMBER 2 Scientists NIH Budget: A Moving Target Adapting to $1.629 billion: Amount Republicans in the biomedical research. To join, go to www.ascb. Change U.S. House of Representatives want to cut from org/Project50. Page 3 the U.S. National Institutes of n Become a member of the Health (NIH) FY11 budget Coalition for the Life Sciences’ (compared with FY10) Congressional Liaison Committee Finding the $745 million: Amount U.S. (CLC). Receive alerts on President Obama wants to add important science policy issues, Right Academic to the NIH budget for FY12 help build local support for Career (compared with FY10) biological research, and visit your Page 11 No matter how you look at it, Representative in the House and the news could be better for the Senators. To become a member, NIH budget. While the budget’s go to www.coalitionforlifesciences. Genomics future is uncertain, one thing org/be-an-advocate/about-the-clc/ Revolutionizes isn’t—Congress needs to hear join-the-clc. from you. n Stay informed about science policy by Cell Biology What can you do? Become more involved in reading the ASCB Newsletter’s Public Policy science policy advocacy. Briefings. Page 22 n Join Project 50, the ASCB Public Policy n Stay up-to-date on current advocacy alerts by Advocacy Team. Receive special briefings visiting http://capwiz.com/jscpp/home. n Inside and organize your colleagues in support of —Kevin M. Wilson 2011 Congressional Biomedical Don’t Miss President’s Column 3 Public Policy Briefing 7 Research Caucus Topics iBioMagazine Call for Nominations 9 Each year the Coalition for the Life Sciences (CLS) plans a Issue 3 Dear Labby 10 series of caucuses on Capitol Hill that are designed to foster Join MAC 10 an appreciation for and understanding of biomedical research. -
Appendix Ii –Curricula Vitae
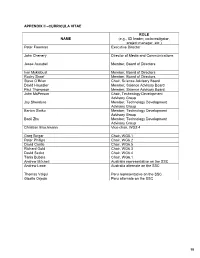
APPENDIX II –CURRICULA VITAE ROL E NAME (e.g., ICI leader, co-investigator, project manager, etc.) Peter Freeman Executive Director John Chenery Director of Media and Communications Jesse Ausubel Member, Board of Directors Ivar Myklebust Member, Board of Directors Rocky Skeef Member, Board of Directors Steve O’Brien Chair, Science Advisory Board David Haussler Member, Science Advisory Board Paul Thompson Member, Science Advisory Board John McPerson Chair, Technology Development Advisory Group Jay Shendure Member, Technology Development Advisory Group Barton Slatko Member, Technology Development Advisory Group Baoli Zhu Member, Technology Development Advisory Group Christian Brochmann Vice-chair, WG2.4 Greg Singer Chair, WG5.1 Peter Phillips Chair, WG6.2 David Castle Chair, WG6.5 Richard Gold Chair, WG6.3 David Secko Chair, WG6.4 Tania Bubela Chair, WG6.1 Andrew Mitchell Australia representative on the SSC Andrew Lowe Australia alternate on the SSC Thomas Valqui Peru representative on the SSC Gisella Orjeda Peru alternate on the SSC 98 PETER FREEMAN BA, PHD, FIBD 120 Stuart Street, Guelph, Ontario, CANADA N1E 4S8 Phone (W): +1 519 824 4120 Cell: +1 519 731 2163 Email: [email protected] PROFILE Experienced science-based (PhD qualified) executive director, with a track record of successfully co- ordinating large multi-institutional research projects, networks and consortia in genomics, proteomics, stem cell research and population health. Strong financial management, strategic planning and project management skills gained in R&D and operations roles in the international malting and brewing industry Superior mentoring, facilitating, technical writing and editing skills used to develop successful interdisciplinary funding proposals. Fully proficient in information /communication technologies and their use in knowledge translation and public outreach activities. -

Academy Honors 17 for Major Contributions to Science
Proc. Natl. Acad. Sci. USA Vol. 96, pp. 1173–1174, February 1999 From the Academy Academy Honors 17 for Major Contributions to Science The National Academy of Sciences has selected 17 in electricity, magnetism, or radiant energy, broadly individuals to receive awards honoring their outstanding interpreted—goes to John Clarke, Luis W. Alvarez contributions to science. The awards will be presented Memorial Chair for Experimental Physics and professor on April 26 at a ceremony in Washington, DC, during the of physics, University of California, Berkeley. Clarke Academy’s 136th annual meeting. Awards and recipients was chosen “for his major contributions to the develop- are as follows: ment of superconducting quantum interference devices (SQUIDS) and their use for scientific measurements, Arctowski Medal—a prize of $20,000, plus $60,000 to an especially involving electricity, magnetism, and electro- institution of the recipient’s choice, awarded every three magnetic waves.” The prize was established through the years to further research in solar physics and solar- Cyrus B. Comstock Fund and has been presented since terrestrial relationships—goes to Arthur J. Hund- 1913. hausen, senior scientist emeritus, High Altitude Obser- vatory, National Center for Atmospheric Research, The Arthur L. Day Prize and Lectureship—a prize of Boulder. Hundhausen was chosen “for his exceptional $20,000 awarded approximately every three years to a research in solar and solar-wind physics, particularly in scientist making new contributions to the physics of the the area of coronal and solar-wind disturbances.” The Earth and whose four to six lectures would prove a solid, medal was established in honor of Henryk Arctowski timely, and useful addition to the knowledge and liter- and has been awarded since 1969. -

Celebrating Years
Chestnut Hill College Department of Biology Celebrating Biomedical Lecture Series 20 Distinguished Speakers Years 1994 – 2013 Keynote Speaker Keynote Speaker 10th Anniversary 20th Anniversary March 2004 September 2013 Stanley B. Prusiner, M.D. Michael S. Brown, M.D. Nobel Prize for Medicine Nobel Prize for Medicine or Physiology or Physiology ii GOALS AND VISION Of the BIOmeDIcAL Lecture SerIeS GOALS AND VISION v To provide a forum at Chestnut Hill College for learning Office of the President recent advances in biomedical research. Since 1994, Chestnut Hill College has been enhanced and broadened by the Biomedical To expose our students and faculty to prominent Seminar Series sponsored by the Biology Department. This series presents students with an v opportunity to meet, interact, listen, and learn from some of the most prestigious scientists in the scientific and medical professionals. country. Over the years, these encounters have opened the doors of research laboratories to our students to provide them with stellar internships and incomparable mentors. It is no exaggeration to say that through these encounters lives have been changed, careers launched, v To foster meaningful interactions and collaboration and horizons expanded. among our neighboring institutions. This program of lectures is genuinely unique; unique because, unlike similar series at large universities, it is neither endowed nor funded by an external source. On the contrary, prominent scientists spend a day of their precious time with Chestnut Hill students out of the To develop new contacts for all our students who are generous bounty of their hearts. Among them are Presidents of institutions, two Nobel v Laureates, Directors of Centers, Senior Administrators, elected members of the National interested in pursuing careers in biotechnology, basic Academy of Sciences, Department Chairpersons, Distinguished Scholars, and Lasker Award recipients. -

Tishkoff SA CV Bibliography.09.08.20.Web
Curriculum Vitae Sarah A. Tishkoff, Ph.D. Date: 09/08/20 Address: 428 Clinical Research Building 415 Curie Boulevard Philadelphia, PA 19104-6145 USA [email protected] Citizenship: U.S. citizen Education: 1989 B.S. University of California, Berkeley (Anthropology & Genetics) 1992 M.Phil. Yale University (Human Genetics, Kenneth Kidd adviser) 1996 Ph.D. Yale University (Genetics, Kenneth Kidd adviser) Postgraduate Training and Fellowship Appointments: 1997-2000 Postdoctoral Fellow, Pennsylvania State University, Department of Biology (Andrew Clark adviser) 1997 Visiting Research Fellow, South African Institute of Medical Research, University of the Witwatersrand, Department of Human Genetics (Trefor Jenkins adviser) Faculty Appointments: 2000-2005 Assistant Professor, University of Maryland, Department of Biology 2005-2007 Associate Professor, University of Maryland, Department of Biology 2008-2012 David and Lyn Silfen University Associate Professor, Department of Genetics, Perelman School of Medicine and Department of Biology, School of Arts and Sciences, University of Pennsylvania (Penn Integrates Knowledge Associate Professor) 2012-present David and Lyn Silfen University Professor, Department of Genetics, Perelman School of Medicine and Department of Biology, School of Arts and Sciences, University of Pennsylvania (Penn Integrates Knowledge Professor) Other Appointments: 2008-present Member, Genomics and Computational Biology (GCB) Graduate Group, Biomedical Graduate Studies, Perelman School of Medicine, University of Pennsylvania -

Pdf File Available
S!JE3JECTINDEEX Volumes 1-15 Numbers following each subject entry direct the reader to the volume and then to the first page of the essay in which reference to the subject occurs. Thus, 3:608 refers the reader to page 608 in Vol. 3. Mention or discussion of individuals in an essay often includes refer- ence to one or more of that person’s published papers. Therefore, readers doing name searches should also consult the Cited-Author Index in each volume. A academies of science 3:337, 3:492, 3587,3:675 academy memberships AAPG Bul/etin—American Association of Petro/eum Geologists (journal) Djerassi, Carl 5:727 3:484 Franklin, Benjamin 5:708 Aaronson, S. Koprowski, Hilary 5:617 guest essay, style in scientific writing 3:4 Krebs, Sir Hans 5:630 reprint, citation analysis 2:286 most-cited authors 5:428, 5:536, 5:547, Abbott, Berenice 12:57 5:569 abbreviations in article titles 1:381 Academy of Lynxes 4:395 ABC (American Broadcasting Academy of Natural Sciences 5:361, Companies) 4:471 5:362 Abel Award 3608 Accademia del Lincei 4:395 Abelson, Philip H. 5:256, 10:219,15:61 Accelerated Surface Post (ASP) 4:25 Abercrombie, Michael 12:156 accidents Aborn, Peter 3:666,4:353,5:454,6:31, children 6:396 6:36,7:290 risks of new technologies 5:643 abortion 5:230 accommodations at conferences 3668 and medical genetics 5:231 accuracy in citations 13367 Abraham, Edward P. 12:102 acetaminophen 5:240 Abrikosov, A.A. 11:134 acetylsalicylic acid 5:236 abstract journals 3:405,4523, 10:16500 acid rain abstracting and indexing setvices 8:366, defined and explained 8:77 14:74 public issues and proposed solutions &88 biomedicine 4:522 Ackerman, Diane 6:227 abstracts in ISI products 15:100, 15:110, acne vulgaris 5:364 15:236 acoustics 12302 added to CCOD 14:74 ISI headquarters 4:352 included in CD-ROM 15:236 restaurants 4:308 Abt, Helmut A. -

Yearbook 2004 Yearbook 2004
COVER((OK)) 22-09-2004 12:00 Pagina 1 PONTIFICIA ACADEMIA SCIENTIARVM PONTIFICIA ACADEMIASCIENTIARVM YEARBOOK 2004 YEARBOOK 2004 VATICAN CITY 01_Prima Parte+BIO.A_1.Prima Parte 25/06/10 10:34 Pagina 1 THE PONTIFICAL ACADEMY OF SCIENCES Yearbook 2004 Vatican City 01_Prima Parte+BIO.A_1.Prima Parte 25/06/10 10:34 Pagina 2 ‘Truth, freedom and responsibility are connected in the experience of the sci- entist. In setting out on his path of research, he understands that he must tread not only with the impartiality required by the objectivity of his method but also with the intellectual honesty, the responsibility, and I would say with a kind of “reverence”, which befit the human spirit in its drawing near to truth. For the scientist, to understand in an ever better way the particular reality of man in relation to the biological-physical processes of nature, to discover always new aspects of the cosmos, to know more about the location and the distri- bution of resources, the social and environmental dynamics, and the logic of progress and development, becomes translated into a duty to serve more fully the whole of mankind, to which he belongs. For this reason, the ethical and moral responsibilities connected to scientific research can be perceived as a requirement within science, because it is a fully human activity, but not as control, or worse, as an imposition which comes from outside. The man of science knows perfectly, from the point of view of his knowledge, that truth cannot be subject to negotiation, cannot be obscured or abandoned to free conventions or agreements between groups of power, societies, or States. -

50Th Anniversary EMBO Members Meeting 2014
WINTER 2014 | 2015 ISSUE 29 50th Anniversary The EMBO Meeting is EMBO Members coming to Birmingham Meeting 2014 page 4 pageS 10 – 11 Meet the scientist Michael Potente pageS 6 – 7 EMBO Young Investigator Investigator EMBO Young News Bringing data to the fore: Interview We need a CERN for Feature Euro-BioImaging: Open access The SourceData project enters genomics: Emmanouil Dermitzakis infrastructure for imaging technologies. proof-of-principle phase talks to Thomas Lemberger. PAGE 2 PAGE 8 PAGE 9 www.embo.org NEWS appointed members of the Scientific Advisory Bringing data to the fore Board will provide us with essential guidance in this respect.” The SourceData project improves access to the data in published scientific The Scientific Advisory Board ofSourceData articles and enables further use of research results. This EMBO initiative includes Jason Swedlow, of the University of has entered its proof-of-principle phase with the support of the Robert Dundee, Scotland, Alfonso Valencia, Director of the Spanish National Bioinformatics Institute, Bosch Foundation. The tools that are being developed as part of the project Susanna-Assunta Sansone, of the University make the source data in scientific publications easier to find and allow of Oxford e-Research Centre, Phil Archer, Data scientists to use the data to their full potential. Activity Lead of the World Wide Web Consortium, and Mark Patterson, Executive Director of eLife. fficient access to primary research could essential findings in the paper to the reader. At The SourceData website will be launched in significantly speed up science. Yet scientific EMBO Press, dedicated scientific data editors April at sourcedata.embo.org Ejournals publish most data as figures that already work with the authors of manuscripts to do not easily allow further analysis of the data optimise and structure the presentation of their and are inaccessible to systematic data mining or figures and data.