Study on Fusion Protein and Its Gene in Baculovirus Specificity Waleed
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Calling and Mating Behavior of Diaphania Angustalis (Lepidoptera: Crambidae)
Copyedited by: OUP Journal of Economic Entomology, XX(X), 2018, 1–5 doi: 10.1093/jee/toy179 Forest Entomology Research Calling and Mating Behavior of Diaphania angustalis (Lepidoptera: Crambidae) Xianhui Shi,1,* Tao Ma,1,2,* Shengnan Zhang,1 Zhaohui Sun,1 Xiaoyang Chen,1 Changlu Wang,3 Caijuan Jia,4 Yongchan Liang,1 Ying Zhu,1 Yurong He,2 and Xiujun Wen1,5 1Guangdong Key Laboratory for Innovative Development and Utilization of Forest Plant Germplasm, College of Forestry and Landscape Architecture, South China Agricultural University, Guangzhou 510642, China, 2College of Agriculture, South China Agricultural University, Guangzhou 510642, China, 3Department of Entomology, Rutgers University, New Brunswick, NJ 08901, 4Wutongshan Park, Shenzhen 518114, China, and 5Corresponding author, e-mail: [email protected] *These authors contributed equally to this work. Subject Editor: Brian Sullivan Received 30 January 2018; Editorial decision 5 June 2018 Abstract Diaphania angustalis Snellen (Lepidoptera: Crambidae) has emerged as a very important pest of blackboard tree, Alstonia scholaris (L.) R. Br. (Apocynaceae), in China during the last two decades. Understanding its biology and behavior is crucial for designing effective and environmentally friendly pest management strategies. Under laboratory conditions [25–28°C, 12:12 (L:D) h, 75–80% RH], adults emerged during both light and dark hours with peak emergence occurring between the first and fifth hours of scotophase, and unmated males and females lived for a mean (±SE) 5.4 ± 0.4 and 5.3 ± 0.7 d, respectively. Female calling behavior was observed only during scotophase (peaking in the fifth hour) by 1- to 5-d-old females, and mating activities occurred 2–5 d after emergence, peaking on day 3. -

Elimination Pathways of Fusion Protein and Peptide Drugs
Review Volume 11, Issue 2, 2021, 9139 - 9147 https://doi.org/10.33263/BRIAC112.91399147 Elimination Pathways of Fusion Protein and Peptide Drugs Ali Khodadoust 1 , Hossein Aghamollaei 2 , Ali Mohammad Latifi 1 , Kazem Hasanpour 3 , Mahdi Kamali 4 , Hamid Tebyanian 5 , Gholamreza Farnoosh 1,* 1 Applied Biotechnology Research Center, Baqiyatallah University of Medical Sciences, Tehran, Iran 2 Chemical Injuries Research Center, Systems biology and Poisonings Institute, Baqiyatallah University of Medical Sciences, Tehran, Iran 3 Sabzevar University of Medical Sciences, School of Medicine, Sabzevar, Iran 4 Nanobiotechnology Research Center, Baqiyatallah University of Medical Sciences, Tehran, Iran 5 Research Center for Prevention of Oral and Dental Diseases, Baqiyatallah University of Medical Sciences, Tehran, Iran * Correspondence: [email protected]; Scopus Author ID 55855454400 Received: 28.07.2020; Revised: 25.08.2020; Accepted: 27.08.2020; Published: 1.09.2020 Abstract: Fusion proteins have been known as an interesting subject for scientific researches in improving properties or making a new function by synergistically incorporating two protein domains into one complex. Fusion proteins are created by ligation of two or more protein domains in a single polypeptide with functional properties. Improvement of therapeutic function is one of the main goals for developing these products. Elimination from the body is one of the most important points that should be considered in the design and production of fusion proteins. This review describes some of the most important excretion/elimination pathways of fusion peptide and protein drugs as well as serious elimination challenges in the development and manufacturing of fusion proteins. Keywords: Fusion proteins; Therapy; Pharmacokinetics; Serum half-life; Peptide. -

UG ETD Template
Characterization of Autographa californica nucleopolyhedrovirus immediate early protein ME53: The role of conserved domains in BV production, viral gene transcription, and evidence for ME53 presence at the ribosome by Robyn Ralph A Thesis presented to The University of Guelph In partial fulfilment of requirements for the degree of Master of Science in Molecular and Cellular Biology Guelph, Ontario, Canada © Robyn Ralph, December, 2018 ABSTRACT CHARACTERIZATION OF AUTOGRAPHA CALIFORNICA NUCLEOPOLYHEDROVIRUS IMMEDIATE EARLY PROTEIN ME53: THE ROLE OF CONSERVED DOMAINS IN BV PRODUCTION, VIRAL GENE TRANSCRIPTION, AND EVIDENCE FOR ME53 PRESENCE AT THE RIBOSOME Robyn Ralph Advisors: University of Guelph, 2018 Dr. Peter Krell Dr. Sarah Wooton The baculovirus AcMNPV early/late gene me53 is required for efficient BV production and is conserved in all alpha and betabaculoviruses. The 449-amino acid protein contains several highly conserved functionally important domains including two putative C4 zinc finger domains (ZnF-N and ZnF-C) whose cysteine residues are 100% conserved. One purpose of this study is to confirm the presence of two zinc binding domains in ME53, as well as determine their role in virus infection and viral gene transcription. Interestingly, deletion of ZnF-C results in an early delay of BV production from 12 to 18 hours post transfection correlating to ME53's cytoplasmic localization. Cytoplasmic functions at early times post-transfection may include translational regulation, which is supported by yeast-2-hybrid data that ME53 interacts with the host 40S ribosomal subunit protein RACK1. In this study the association of ME53 with the ribosomes of virus infected cells was also investigated. iii DEDICATION I dedicate this thesis to my father, Ronald James Ralph. -

Tically Expands Our Understanding on Virosphere in Temperate Forest Ecosystems
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 21 June 2021 doi:10.20944/preprints202106.0526.v1 Review Towards the forest virome: next-generation-sequencing dras- tically expands our understanding on virosphere in temperate forest ecosystems Artemis Rumbou 1,*, Eeva J. Vainio 2 and Carmen Büttner 1 1 Faculty of Life Sciences, Albrecht Daniel Thaer-Institute of Agricultural and Horticultural Sciences, Humboldt-Universität zu Berlin, Ber- lin, Germany; [email protected], [email protected] 2 Natural Resources Institute Finland, Latokartanonkaari 9, 00790, Helsinki, Finland; [email protected] * Correspondence: [email protected] Abstract: Forest health is dependent on the variability of microorganisms interacting with the host tree/holobiont. Symbiotic mi- crobiota and pathogens engage in a permanent interplay, which influences the host. Thanks to the development of NGS technol- ogies, a vast amount of genetic information on the virosphere of temperate forests has been gained the last seven years. To estimate the qualitative/quantitative impact of NGS in forest virology, we have summarized viruses affecting major tree/shrub species and their fungal associates, including fungal plant pathogens, mutualists and saprotrophs. The contribution of NGS methods is ex- tremely significant for forest virology. Reviewed data about viral presence in holobionts, allowed us to address the role of the virome in the holobionts. Genetic variation is a crucial aspect in hologenome, significantly reinforced by horizontal gene transfer among all interacting actors. Through virus-virus interplays synergistic or antagonistic relations may evolve, which may drasti- cally affect the health of the holobiont. Novel insights of these interplays may allow practical applications for forest plant protec- tion based on endophytes and mycovirus biocontrol agents. -

CUE-101, a Novel Fc Fusion Protein for Selective Targeting and Expansion of Anti-Tumor T Cells for Treatment of HPV-Driven Malignancies
CUE-101, a novel Fc fusion protein for selective targeting and expansion of anti-tumor T cells for treatment of HPV-driven malignancies Natasha Girgis1, Steven N. Quayle1, Alyssa Nelson1, Dharma Raj Thapa1, Sandrine Hulot1, Lauren Kraemer1, Miguel Moreta1, Zohra Merazga1, Robert Ruidera1, Dominic Beal1, Mark Haydock1, Jonathan Soriano1, Luke Witt1, Jessica Ryabin1, Emily Spaulding1, John F. Ross1, Saso Cemerski1, Anish Suri1, Rodolfo Chaparro1, Ronald Seidel1, Kenneth J. Pienta2, Mary C. Simcox1 1Cue Biopharma, Cambridge, Massachusetts; 2The James Buchanan Brady Urological Institute and the Department of Urology, Johns Hopkins School of Medicine, Baltimore, Maryland + + Background CUE-101 selectively expands E711-20-specific CD8 T mCUE-101 expands functional antigen-specific CD8 T • Human papilloma virus (HPV) is responsible for 72% of oropharyngeal, 90% of cervical, 90% cells from healthy human PBMCs cells in the tumor and the periphery of anal, and 71% of vulvar, vaginal, or penile cancers, causing significant morbidity and A. B. A. B. C. 9 mortality worldwide. Innovative therapies are urgently needed for these malignancies, Vehicle 100 nM CUE-101 107 15 Peripheral Blood particularly in the largely incurable metastatic setting. 106 10 5 • The E7 oncoprotein is constitutively expressed in HPV-associated cancers, is necessary for 105 ⍺PD-1 5 PE 4 Vehicle mCUE-101 Combo - 10 initiation and maintenance of malignant transformation, and is genetically conserved in 1 cancer (Mirabello 2017). 103 1 1 102 E7-Specific of CD8+ T cells % IFNg+,TNFa+ -

Interferon-Based Biopharmaceuticals: Overview on the Production, Purification, and Formulation
Review Interferon-Based Biopharmaceuticals: Overview on the Production, Purification, and Formulation Leonor S. Castro 1,†, Guilherme S. Lobo 1,†, Patrícia Pereira 2 , Mara G. Freire 1 ,Márcia C. Neves 1,* and Augusto Q. Pedro 1,* 1 CICECO–Aveiro Institute of Materials, Chemistry Department, University of Aveiro, Campus Universitário de Santiago, 3810-193 Aveiro, Portugal; [email protected] (L.S.C.); [email protected] (G.S.L.); [email protected] (M.G.F.) 2 Centre for Mechanical Engineering, Materials and Processes, Department of Chemical Engineering, University of Coimbra, Rua Sílvio Lima-Polo II, 3030-790 Coimbra, Portugal; [email protected] * Correspondence: [email protected] (M.C.N.); [email protected] (A.Q.P.) † These authors contributed equally to this work. Abstract: The advent of biopharmaceuticals in modern medicine brought enormous benefits to the treatment of numerous human diseases and improved the well-being of many people worldwide. First introduced in the market in the early 1980s, the number of approved biopharmaceutical products has been steadily increasing, with therapeutic proteins, antibodies, and their derivatives accounting for most of the generated revenues. The success of pharmaceutical biotechnology is closely linked with remarkable developments in DNA recombinant technology, which has enabled the production of proteins with high specificity. Among promising biopharmaceuticals are interferons, first described by Isaacs and Lindenmann in 1957 and approved for clinical use in humans nearly thirty years later. Interferons are secreted autocrine and paracrine proteins, which by regulating several biochemical Citation: Castro, L.S.; Lobo, G.S.; pathways have a spectrum of clinical effectiveness against viral infections, malignant diseases, Pereira, P.; Freire, M.G.; Neves, M.C.; and multiple sclerosis. -
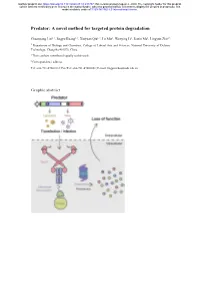
A Novel Method for Targeted Protein Degradation
bioRxiv preprint doi: https://doi.org/10.1101/2020.07.31.231787; this version posted August 2, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Predator: A novel method for targeted protein degradation Chuanyang Liu1, †, Jingyu Kuang1, †, Xinyuan Qiu1, †, Lu Min1, Wenying Li1, Jiaxin Ma1, Lingyun Zhu1,* 1 Department of Biology and Chemistry, College of Liberal Arts and Sciences, National University of Defense Technology, Changsha 410073, China † These authors contributed equally to this work. ∗ Correspondence address. Tel: +86-731-87001812; Fax/Tel: +86-731-87001801; E-mail: [email protected] Graphic abstract bioRxiv preprint doi: https://doi.org/10.1101/2020.07.31.231787; this version posted August 2, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abstract Protein expression and degradation are fundamental to cell function and physiological status of organisms. Interfering with protein expression not only provides powerful strategies to analyze the function of proteins but also inspires effective treatment methods for diseases caused by protein dysfunction. Recently, harnessing the power of the ubiquitin-proteasome system for targeted protein degradation (TPD) has become the focus of researches. Over the past two decades, TPD technologies, such as E3 ligase modification, PROTACs, and the Trim-Away method, have successfully re-oriented the ubiquitin-proteasome pathway and thus degraded many pathogenic proteins and even "undruggable" targets. -

Overview of Antibody Drug Delivery
pharmaceutics Review Overview of Antibody Drug Delivery Sahar Awwad 1,2,* ID and Ukrit Angkawinitwong 1 1 UCL School of Pharmacy, London WC1N 1AX, UK; [email protected] 2 National Institute for Health Research (NIHR) Biomedical Research Centre at Moorfields Eye Hospital NHS Foundation Trust and UCL Institute of Ophthalmology, London EC1 V9EL, UK * Correspondence: [email protected]; Tel.: +44-207-753-5802 Received: 27 March 2018; Accepted: 29 June 2018; Published: 4 July 2018 Abstract: Monoclonal antibodies (mAbs) are one of the most important classes of therapeutic proteins, which are used to treat a wide number of diseases (e.g., oncology, inflammation and autoimmune diseases). Monoclonal antibody technologies are continuing to evolve to develop medicines with increasingly improved safety profiles, with the identification of new drug targets being one key barrier for new antibody development. There are many opportunities for developing antibody formulations for better patient compliance, cost savings and lifecycle management, e.g., subcutaneous formulations. However, mAb-based medicines also have limitations that impact their clinical use; the most prominent challenges are their short pharmacokinetic properties and stability issues during manufacturing, transport and storage that can lead to aggregation and protein denaturation. The development of long acting protein formulations must maintain protein stability and be able to deliver a large enough dose over a prolonged period. Many strategies are being pursued to improve the formulation and dosage forms of antibodies to improve efficacy and to increase the range of applications for the clinical use of mAbs. Keywords: antibodies; protein; pharmacokinetics; drug delivery; stability 1. -

Insect Pathogens As Biological Control Agents: Back to the Future ⇑ L.A
Journal of Invertebrate Pathology 132 (2015) 1–41 Contents lists available at ScienceDirect Journal of Invertebrate Pathology journal homepage: www.elsevier.com/locate/jip Insect pathogens as biological control agents: Back to the future ⇑ L.A. Lacey a, , D. Grzywacz b, D.I. Shapiro-Ilan c, R. Frutos d, M. Brownbridge e, M.S. Goettel f a IP Consulting International, Yakima, WA, USA b Agriculture Health and Environment Department, Natural Resources Institute, University of Greenwich, Chatham Maritime, Kent ME4 4TB, UK c U.S. Department of Agriculture, Agricultural Research Service, 21 Dunbar Rd., Byron, GA 31008, USA d University of Montpellier 2, UMR 5236 Centre d’Etudes des agents Pathogènes et Biotechnologies pour la Santé (CPBS), UM1-UM2-CNRS, 1919 Route de Mendes, Montpellier, France e Vineland Research and Innovation Centre, 4890 Victoria Avenue North, Box 4000, Vineland Station, Ontario L0R 2E0, Canada f Agriculture and Agri-Food Canada, Lethbridge Research Centre, Lethbridge, Alberta, Canada1 article info abstract Article history: The development and use of entomopathogens as classical, conservation and augmentative biological Received 24 March 2015 control agents have included a number of successes and some setbacks in the past 15 years. In this forum Accepted 17 July 2015 paper we present current information on development, use and future directions of insect-specific Available online 27 July 2015 viruses, bacteria, fungi and nematodes as components of integrated pest management strategies for con- trol of arthropod pests of crops, forests, urban habitats, and insects of medical and veterinary importance. Keywords: Insect pathogenic viruses are a fruitful source of microbial control agents (MCAs), particularly for the con- Microbial control trol of lepidopteran pests. -

COVID-19: Mechanisms of Vaccination and Immunity
Review COVID-19: Mechanisms of Vaccination and Immunity Daniel E. Speiser 1,* and Martin F. Bachmann 2,3,4,* 1 Department of Oncology, University Hospital and University of Lausanne, 1066 Lausanne, Switzerland 2 International Immunology Centre, Anhui Agricultural University, Hefei 230036, China 3 Department of Rheumatology, Immunology and Allergology, Inselspital, University of Bern, 3010 Bern, Switzerland 4 Department of BioMedical Research, University of Bern, 3008 Bern, Switzerland * Correspondence: [email protected] (D.E.S.); [email protected] (M.F.B.) Received: 2 July 2020; Accepted: 20 July 2020; Published: 22 July 2020 Abstract: Vaccines are needed to protect from SARS-CoV-2, the virus causing COVID-19. Vaccines that induce large quantities of high affinity virus-neutralizing antibodies may optimally prevent infection and avoid unfavorable effects. Vaccination trials require precise clinical management, complemented with detailed evaluation of safety and immune responses. Here, we review the pros and cons of available vaccine platforms and options to accelerate vaccine development towards the safe immunization of the world’s population against SARS-CoV-2. Favorable vaccines, used in well-designed vaccination strategies, may be critical for limiting harm and promoting trust and a long-term return to normal public life and economy. Keywords: SARS-CoV-2; COVID-19; nucleic acid tests; serology; vaccination; immunity 1. Introduction The COVID-19 pandemic holds great challenges for which the world is only partially prepared [1]. SARS-CoV-2 combines serious pathogenicity with high infectivity. The latter is enhanced by the fact that asymptomatic and pre-symptomatic individuals can transmit the virus, in contrast to SARS-CoV-1 and MERS-CoV, which were transmitted by symptomatic patients and could be contained more efficiently [2,3]. -

Incubation Period of Measles from Exposure to Prodrome ■ Exposure to Rash Onset Averages 11 to 12 Days
Measles Paul Gastanaduy, MD; Penina Haber, MPH; Paul A. Rota, PhD; and Manisha Patel, MD, MS Measles is an acute, viral, infectious disease. References to measles can be found from as early as the 7th century. The Measles disease was described by the Persian physician Rhazes in the ● Acute viral infectious disease 10th century as “more to be dreaded than smallpox.” ● First described in 7th century In 1846, Peter Panum described the incubation period of ● Vaccines first licensed include measles and lifelong immunity after recovery from the disease. measles in 1963, MMR in 1971, John Enders and Thomas Chalmers Peebles isolated the virus in and MMRV in 2005 human and monkey kidney tissue culture in 1954. The first live, ● Infection nearly universal attenuated vaccine (Edmonston B strain) was licensed for use in during childhood in the United States in 1963. In 1971, a combined measles, mumps, prevaccine era and rubella (MMR) vaccine was licensed for use in the United ● Still common and often fatal States. In 2005, a combination measles, mumps, rubella, and in developing countries varicella (MMRV) vaccine was licensed. Before a vaccine was available, infection with measles virus was nearly universal during childhood, and more than 90% of persons were immune due to past infection by age 15 years. Measles is still a common and often fatal disease in developing countries. The World Health Organization estimates there were 142,300 deaths from measles globally in 2018. In the United 13 States, there have been recent outbreaks; the largest occurring in 2019 primarily among people who were not vaccinated. -

Thermal Requirements of Trichogramma
Revista Colombiana de Entomología 2021, 47 (1): e8548 • https://doi.org/10.25100/socolen.v47i1.8548 Sección Agrícola / Agriculture Artículo de investigación / Research paper Thermal requirements of Trichogramma pretiosum (Hymenoptera: Trichogrammatidae) lines in Neoleucinodes elegantalis (Lepidoptera: Crambidae) eggs Requerimientos térmicos de las líneas de Trichogramma pretiosum (Hymenoptera: Trichogrammatidae) en huevos de Neoleucinodes elegantalis (Lepidoptera: Crambidae) CRISTIANE RAMOS COUTINHO1; SUYANNE ARAÚJO DE SOUZA2; ANTÔNIA DÉBORA DOS SANTOS PONTES3; MAURICIO SEKIGUCHI DE GODOY4; FABRICIO FAGUNDES PEREIRA5; PATRIK LUIZ PASTORI6 1 Ph. D. in Agronomy/Entomology, IN Soluções Biológicas LTDA., Fortaleza, Ceará State, Brazil, [email protected], https://orcid. org/0000-0002-7870-904X. 2 Engineer Agronomist. Universidade Estadual Paulista “Julio de Mesquita Filho” - UNESP, Faculdade de Ciências Agronômi- cas, Câmpus de Botucatu, Botucatu, São Paulo State, Brazil, [email protected], https://orcid.org/0000-0001-5524-6989. 3 Student in Agronomy. Universidade Federal do Ceará - UFC, Departamento de Fitotecnia, Fortaleza, Ceará State, Brazil, [email protected], https://orcid.org/0000-0001- 5299-2783. 4 Ph. D. in Agronomy/Entomology, Universidade Federal Rural do Semi-Árido - UFERSA, Centro de Ciências Agrárias, Departamento de Ciências Agronômicas e Florestais, Mossoró, Rio Grande do Norte State, Brazil, [email protected], https://orcid.org/0000-0002-3842-340X. 5 Ph. D. in Ento- mology, Universidade Federal da Grande Dourados - UFGD, Faculdade de Ciências Biológicas e Ambientais, Rodovia Dourados/Itahum, Dourados, Mato Grosso do Sul State, Brazil, [email protected], https://orcid.org/0000-0003-1638-7409. 6 Ph. D. in Agronomy/Entomology. Universidade Federal do Ceará - UFC, Departamento de Fitotecnia, Fortaleza, Ceará State, Brazil, [email protected], https://orcid.org/0000-0003-1892-8029.