Horizontal Gene Transfer Contributes to the Evolution of Arthropod Herbivory
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Review of Sampling and Monitoring Methods for Beneficial Arthropods
insects Review A Review of Sampling and Monitoring Methods for Beneficial Arthropods in Agroecosystems Kenneth W. McCravy Department of Biological Sciences, Western Illinois University, 1 University Circle, Macomb, IL 61455, USA; [email protected]; Tel.: +1-309-298-2160 Received: 12 September 2018; Accepted: 19 November 2018; Published: 23 November 2018 Abstract: Beneficial arthropods provide many important ecosystem services. In agroecosystems, pollination and control of crop pests provide benefits worth billions of dollars annually. Effective sampling and monitoring of these beneficial arthropods is essential for ensuring their short- and long-term viability and effectiveness. There are numerous methods available for sampling beneficial arthropods in a variety of habitats, and these methods can vary in efficiency and effectiveness. In this paper I review active and passive sampling methods for non-Apis bees and arthropod natural enemies of agricultural pests, including methods for sampling flying insects, arthropods on vegetation and in soil and litter environments, and estimation of predation and parasitism rates. Sample sizes, lethal sampling, and the potential usefulness of bycatch are also discussed. Keywords: sampling methodology; bee monitoring; beneficial arthropods; natural enemy monitoring; vane traps; Malaise traps; bowl traps; pitfall traps; insect netting; epigeic arthropod sampling 1. Introduction To sustainably use the Earth’s resources for our benefit, it is essential that we understand the ecology of human-altered systems and the organisms that inhabit them. Agroecosystems include agricultural activities plus living and nonliving components that interact with these activities in a variety of ways. Beneficial arthropods, such as pollinators of crops and natural enemies of arthropod pests and weeds, play important roles in the economic and ecological success of agroecosystems. -

To Us Insectometers, It Is Clear That Insect Decline in Our Costa Rican
SPECIAL FEATURE: PERSPECTIVE To us insectometers, it is clear that insect decline in our Costa Rican tropics is real, so let’sbekindto SPECIAL FEATURE: PERSPECTIVE the survivors Daniel H. Janzena,1 and Winnie Hallwachsa Edited by David L. Wagner, University of Connecticut, and accepted by Editorial Board Member May R. Berenbaum November 11, 2020 (received for review April 6, 2020) We have been field observers of tropical insects on four continents and, since 1978, intense observers of caterpillars, their parasites, and their associates in the 1,260 km2 of dry, cloud, and rain forests of Area´ de Conservaci ´onGuanacaste (ACG) in northwestern Costa Rica. ACG’s natural ecosystem restoration began with its national park designation in 1971. As human biomonitors, or “insectometers,” we see that ACG’s insect species richness and density have gradually declined since the late 1970s, and more intensely since about 2005. The overarching perturbation is climate change. It has caused increasing ambient tempera- tures for all ecosystems; more erratic seasonal cues; reduced, erratic, and asynchronous rainfall; heated air masses sliding up the volcanoes and burning off the cloud forest; and dwindling biodiversity in all ACG terrestrial ecosystems. What then is the next step as climate change descends on ACG’s many small-scale successes in sustainable biodevelopment? Be kind to the survivors by stimulating and facilitating their owner societies to value them as legitimate members of a green sustainable nation. Encourage national bioliteracy, BioAlfa. climate change | BioAlfa | conservation by rewilding | biodevelopment | insect decline As “insectometers,” also known as human biomoni- eventually focused into Area´ de Conservaci ´onGuana- tors, we have watched the conspicuous ongoing de- caste (ACG) in northwestern Costa Rica (8–13) (Fig. -

4 Reproductive Biology of Cerambycids
4 Reproductive Biology of Cerambycids Lawrence M. Hanks University of Illinois at Urbana-Champaign Urbana, Illinois Qiao Wang Massey University Palmerston North, New Zealand CONTENTS 4.1 Introduction .................................................................................................................................. 133 4.2 Phenology of Adults ..................................................................................................................... 134 4.3 Diet of Adults ............................................................................................................................... 138 4.4 Location of Host Plants and Mates .............................................................................................. 138 4.5 Recognition of Mates ................................................................................................................... 140 4.6 Copulation .................................................................................................................................... 141 4.7 Larval Host Plants, Oviposition Behavior, and Larval Development .......................................... 142 4.8 Mating Strategy ............................................................................................................................ 144 4.9 Conclusion .................................................................................................................................... 148 Acknowledgments ................................................................................................................................. -

Stable Isotope Methods in Biological and Ecological Studies of Arthropods
eea_572.fm Page 3 Tuesday, June 12, 2007 4:17 PM DOI: 10.1111/j.1570-7458.2007.00572.x Blackwell Publishing Ltd MINI REVIEW Stable isotope methods in biological and ecological studies of arthropods CORE Rebecca Hood-Nowotny1* & Bart G. J. Knols1,2 Metadata, citation and similar papers at core.ac.uk Provided by Wageningen University & Research Publications 1International Atomic Energy Agency (IAEA), Agency’s Laboratories Seibersdorf, A-2444 Seibersdorf, Austria, 2Laboratory of Entomology, Wageningen University and Research Centre, P.O. Box 8031, 6700 EH Wageningen, The Netherlands Accepted: 13 February 2007 Key words: marking, labelling, enrichment, natural abundance, resource turnover, 13-carbon, 15-nitrogen, 18-oxygen, deuterium, mass spectrometry Abstract This is an eclectic review and analysis of contemporary and promising stable isotope methodologies to study the biology and ecology of arthropods. It is augmented with literature from other disciplines, indicative of the potential for knowledge transfer. It is demonstrated that stable isotopes can be used to understand fundamental processes in the biology and ecology of arthropods, which range from nutrition and resource allocation to dispersal, food-web structure, predation, etc. It is concluded that falling costs and reduced complexity of isotope analysis, besides the emergence of new analytical methods, are likely to improve access to isotope technology for arthropod studies still further. Stable isotopes pose no environmental threat and do not change the chemistry or biology of the target organism or system. These therefore represent ideal tracers for field and ecophysiological studies, thereby avoiding reductionist experimentation and encouraging more holistic approaches. Con- sidering (i) the ease with which insects and other arthropods can be marked, (ii) minimal impact of the label on their behaviour, physiology, and ecology, and (iii) environmental safety, we advocate more widespread application of stable isotope technology in arthropod studies and present a variety of potential uses. -

Transcriptome and Gene Expression Analysis of Rhynchophorus Ferrugineus (Coleoptera: Curculionidae) During Developmental Stages
Transcriptome and gene expression analysis of Rhynchophorus ferrugineus (Coleoptera: Curculionidae) during developmental stages Hongjun Yang1,2, Danping Xu1, Zhihang Zhuo1,2,3, Jiameng Hu2 and Baoqian Lu4 1 College of Life Science, China West Normal University, Nanchong, Sichuan, China 2 Key Laboratory of Genetics and Germplasm Innovation of Tropical Special Forest Trees and Ornamental Plants, Ministry of Education, Key Laboratory of Germplasm Resources Biology of Tropical Special Ornamental Plants of Hainan Province, College of Forestry, Hainan University, Haikou, Hainan, China 3 Key Laboratory of Integrated Pest Management on Crops in South China, Ministry of Agriculture, South China Agricultural University, Guangzhou, Guangdong, China 4 Key Laboratory of Integrated Pest Management on Tropical Crops, Ministry of Agriculture China, Environ- ment and Plant Protection Institute, Chinese Academy of Tropical Agricultural Sciences, Haikou, Hainan, China ABSTRACT Background. Red palm weevil, Rhynchophorus ferrugineus Olivier, is one of the most destructive pests harming palm trees. However, genomic resources for R. ferrugineus are still lacking, limiting the ability to discover molecular and genetic means of pest control. Methods. In this study, PacBio Iso-Seq and Illumina RNA-seq were used to generate transcriptome from three developmental stages of R. ferrugineus (pupa, 7th-instar larva, adult) to increase the understanding of the life cycle and molecular characteristics of the pest. Results. Sequencing generated 625,983,256 clean reads, from which 63,801 full-length transcripts were assembled with N50 of 3,547 bp. Expression analyses revealed 8,583 differentially expressed genes (DEGs). Moreover, gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis revealed that these Submitted 5 March 2020 Accepted 29 September 2020 DEGs were mainly related to the peroxisome pathway which associated with metabolic Published 2 November 2020 pathways, material transportation and organ tissue formation. -

Feeding Damage by Larvae of the Mustard Leaf Beetle Deters Conspecific Females from Oviposition and Feeding
Chapter 5 Feeding damage by larvae of the mustard leaf beetle deters conspecific females from oviposition and feeding Key words: Chinese cabbage, damage-induc ed response, host acceptance, larval frass, larval performance, larval secretion, oviposition behaviour, Phaedon cochleariae, regurgitant Abstract Herbivorous insects may be informed about the presence of competitors on the same host plant by a variety of cues. These cues may derive from either the competitor itself or the damaged plant. In the mustard leaf beetle Phaedon cochleariae (Coleoptera, Chrysomelidae), adults are known to be deterred from feeding and oviposition by the exocrine glandular secretion of conspecific co-occurring larvae. We hypothesised that the exocrine larval secretion released by feeding larvae may adsorb to the surface of Chinese cabbage leaves, and thus, convey the information about their former or actual presence. Further experiments tested the influence of leaves damaged by conspecific larvae, mechanically damaged leaves, larval frass and regurgitant on the oviposition and feeding behaviour of P. cochleariae. Finally, the effect of previous conspecific herbivory on larval development and larval host selection was assessed. Our results show that (epi)chrysomelidial, the major component of the exocrine secretion from P. cochleariae larvae, was detectable by GC-MS in surface extracts from leaves upon which larvae had fed. However, leaves exposed to volatiles of the larval secretion were not avoided by female P. cochleariae for feeding or oviposition. Thus, we conclude that secretion volatiles did not adsorb in sufficient amounts on the leaf surface to display deterrent activity towards adults. By contrast, gravid females avoided to feed and lay their eggs on leaves damaged by second-instar larvae for 3 d when compared to undamaged leaves. -

Phaedon Desotonis Balsbaugh (Coleoptera: Chrysomelidae), a Coreopsis (Asteaceae) Pest New to Florida
DACS-P-01670 Florida Department of Agriculture and Consumer Services, Division of Plant Industry Charles H. Bronson, Commissioner of Agriculture Phaedon desotonis Balsbaugh (Coleoptera: Chrysomelidae), a Coreopsis (Asteaceae) pest new to Florida Michael C. Thomas, [email protected], Taxonomic Entomologist, Florida Department of Agriculture & Consumer Services, Division of Plant Industry INTRODUCTION: Until 2001, Phaedon desotonis Balsbaugh was known from a single specimen collected in northern Alabama (Balsbaugh and Hays 1972; Balsbaugh 1983). Since then, P. desotonis has been discovered to have a broad distribution in the southeastern United States (Wheeler and Hoebeke 2001) and has emerged as an occasional pest of ornamental plantings of tickseed, Coreopsis spp. (Braman et al 2002), Florida’s official state wildflower. This publication records its presence for the first time in Florida and summarizes the available information on its habits, life history, and pest potential. IDENTIFICATION: The genus Phaedon includes eight described species in the U.S. (Balsbaugh 1983). They are oblong, convex, metallic beetles about 3-5 mm in length. There are only two species known to occur in Florida: the newly recorded P. desotonis (Fig. 1) and the widespread P. viridis (Melsheimer). Phaedon desotonis (Fig. 1) is more elongate, has a greenish pronotum and purplish black elytra, while P. viridis (Fig. 2) is less elongate, and in Florida is entirely bronze. Elsewhere, it may be greenish or bluish. In P. viridis, the anterior borders of the mesosternum and first visible abdominal sternite have very large punctures, while those of P. desotonis do not. The structure of the male genitalia also differs in the two species (see Wheeler and Hoebeke 2001, Fig. -
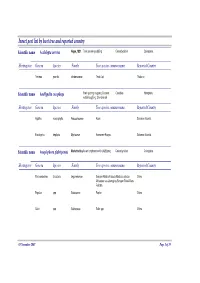
Insect Pest List by Host Tree and Reported Country
Insect pest list by host tree and reported country Scientific name Acalolepta cervina Hope, 1831 Teak canker grub|Eng Cerambycidae Coleoptera Hosting tree Genera Species Family Tree species common name Reported Country Tectona grandis Verbenaceae Teak-Jati Thailand Scientific name Amblypelta cocophaga Fruit spotting bug|eng Coconut Coreidae Hemiptera nutfall bug|Eng, Chinche del Hosting tree Genera Species Family Tree species common name Reported Country Agathis macrophylla Araucariaceae Kauri Solomon Islands Eucalyptus deglupta Myrtaceae Kamarere-Bagras Solomon Islands Scientific name Anoplophora glabripennis Motschulsky Asian longhorn beetle (ALB)|eng Cerambycidae Coleoptera Hosting tree Genera Species Family Tree species common name Reported Country Paraserianthes falcataria Leguminosae Sengon-Albizia-Falcata-Molucca albizia- China Moluccac sau-Jeungjing-Sengon-Batai-Mara- Falcata Populus spp. Salicaceae Poplar China Salix spp. Salicaceae Salix spp. China 05 November 2007 Page 1 of 35 Scientific name Aonidiella orientalis Newstead, Oriental scale|eng Diaspididae Homoptera 1894 Hosting tree Genera Species Family Tree species common name Reported Country Lovoa swynnertonii Meliaceae East African walnut Cameroon Azadirachta indica Meliaceae Melia indica-Neem Nigeria Scientific name Apethymus abdominalis Lepeletier, Tenthredinidae Hymenoptera 1823 Hosting tree Genera Species Family Tree species common name Reported Country Other Coniferous Other Coniferous Romania Scientific name Apriona germari Hope 1831 Long-horned beetle|eng Cerambycidae -

The Molecular Basis for the Neofunctionalization of the Juvenile Hormone Esterase Duplication in Drosophila T
Insect Biochemistry and Molecular Biology 106 (2019) 10–18 Contents lists available at ScienceDirect Insect Biochemistry and Molecular Biology journal homepage: www.elsevier.com/locate/ibmb The molecular basis for the neofunctionalization of the juvenile hormone esterase duplication in Drosophila T ∗ Davis H. Hopkinsa,b, , Rahul V. Raneb, Faisal Younusa,b, Chris W. Coppinb, Gunjan Pandeyb, Colin J. Jacksona, John G. Oakeshottb a Research School of Chemistry, Australian National University, Canberra, Australian Capital Territory, 2601, Australia b CSIRO Land and Water, Black Mountain, Canberra, Australian Capital Territory, 2601, Australia ARTICLE INFO ABSTRACT Keywords: The Drosophila melanogaster enzymes juvenile hormone esterase (DmJHE) and its duplicate, DmJHEdup, present Neofunctionalization ideal examples for studying the structural changes involved in the neofunctionalization of enzyme duplicates. Structural evolution DmJHE is a hormone esterase with precise regulation and highly specific activity for its substrate, juvenile Juvenile hormone esterase hormone. DmJHEdup is an odorant degrading esterase (ODE) responsible for processing various kairomones in Odorant degrading enzyme antennae. Our phylogenetic analysis shows that the JHE lineage predates the hemi/holometabolan split and that several duplications of JHEs have been templates for the evolution of secreted β-esterases such as ODEs through the course of insect evolution. Our biochemical comparisons further show that DmJHE has sufficient substrate promiscuity and activity against odorant esters for a duplicate to evolve a general ODE function against a range of mid-long chain food esters, as is shown in DmJHEdup. This substrate range complements that of the only other general ODE known in this species, Esterase 6. Homology models of DmJHE and DmJHEdup enabled compar- isons between each enzyme and the known structures of a lepidopteran JHE and Esterase 6. -

I, Robot, Can Do That! (Adapted from the 2005 Lost City Expedition)
Thunder Bay Sinkholes 2008 I, Robot, Can Do That! (adapted from the 2005 Lost City Expedition) FOCUS SEATIN G ARRAN G E M ENT Underwater robotic vehicles for scientific explora- Seven groups of students tion MAXI M U M NU M BER O F STUDENTS GRADE LEVE L 35 7-8 (Physical Science/Life Science) KEY WORDS FOCUS QUESTION ABE How can underwater robots be used to assist sci- ROPOS entific explorations? Remotely Operated Vehicle Hercules LEARNIN G OBJECTIVES Tiburon Students will be able to describe and contrast at RCV-150 least three types of underwater robots used for Robot scientific explorations. BAC kg ROUND IN F OR M ATION Students will be able to discuss the advantages In June, 2001, the Ocean Explorer Thunder Bay and disadvantages of using underwater robots in ECHO Expedition was searching for shipwrecks scientific explorations. in the deep waters of the Thunder Bay National Marine Sanctuary and Underwater Preserve in Given a specific exploration task, students will Lake Huron. But the explorers discovered more be able to identify robotic vehicles best suited to than shipwrecks: dozens of underwater sinkholes carry out this task. in the limestone bedrock, some of which were several hundred meters across and 20 meters MATERIA L S deep. The following year, an expedition to sur- Copies of the “Underwater Robot Capability vey the sinkholes found that some of them were Survey,” one for each student group releasing fluids that produced a visible cloudy layer above the lake bottom, and the lake floor AUDIOVISUA L MATERIA L S near some of the sinkholes was covered by con- (Optional) Computers with internet access spicuous green, purple, white, and brown mats. -

Thesis (PDF, 13.51MB)
Insects and their endosymbionts: phylogenetics and evolutionary rates Daej A Kh A M Arab The University of Sydney Faculty of Science 2021 A thesis submitted in fulfilment of the requirements for the degree of Doctor of Philosophy Authorship contribution statement During my doctoral candidature I published as first-author or co-author three stand-alone papers in peer-reviewed, internationally recognised journals. These publications form the three research chapters of this thesis in accordance with The University of Sydney’s policy for doctoral theses. These chapters are linked by the use of the latest phylogenetic and molecular evolutionary techniques for analysing obligate mutualistic endosymbionts and their host mitochondrial genomes to shed light on the evolutionary history of the two partners. Therefore, there is inevitably some repetition between chapters, as they share common themes. In the general introduction and discussion, I use the singular “I” as I am the sole author of these chapters. All other chapters are co-authored and therefore the plural “we” is used, including appendices belonging to these chapters. Part of chapter 2 has been published as: Bourguignon, T., Tang, Q., Ho, S.Y., Juna, F., Wang, Z., Arab, D.A., Cameron, S.L., Walker, J., Rentz, D., Evans, T.A. and Lo, N., 2018. Transoceanic dispersal and plate tectonics shaped global cockroach distributions: evidence from mitochondrial phylogenomics. Molecular Biology and Evolution, 35(4), pp.970-983. The chapter was reformatted to include additional data and analyses that I undertook towards this paper. My role was in the paper was to sequence samples, assemble mitochondrial genomes, perform phylogenetic analyses, and contribute to the writing of the manuscript. -

Phylogenetic and Population Genetic Studies on Some Insect and Plant Associated Nematodes
PHYLOGENETIC AND POPULATION GENETIC STUDIES ON SOME INSECT AND PLANT ASSOCIATED NEMATODES DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Amr T. M. Saeb, M.S. * * * * * The Ohio State University 2006 Dissertation Committee: Professor Parwinder S. Grewal, Adviser Professor Sally A. Miller Professor Sophien Kamoun Professor Michael A. Ellis Approved by Adviser Plant Pathology Graduate Program Abstract: Throughout the evolutionary time, nine families of nematodes have been found to have close associations with insects. These nematodes either have a passive relationship with their insect hosts and use it as a vector to reach their primary hosts or they attack and invade their insect partners then kill, sterilize or alter their development. In this work I used the internal transcribed spacer 1 of ribosomal DNA (ITS1-rDNA) and the mitochondrial genes cytochrome oxidase subunit I (cox1) and NADH dehydrogenase subunit 4 (nd4) genes to investigate genetic diversity and phylogeny of six species of the entomopathogenic nematode Heterorhabditis. Generally, cox1 sequences showed higher levels of genetic variation, larger number of phylogenetically informative characters, more variable sites and more reliable parsimony trees compared to ITS1-rDNA and nd4. The ITS1-rDNA phylogenetic trees suggested the division of the unknown isolates into two major phylogenetic groups: the HP88 group and the Oswego group. All cox1 based phylogenetic trees agreed for the division of unknown isolates into three phylogenetic groups: KMD10 and GPS5 and the HP88 group containing the remaining 11 isolates. KMD10, GPS5 represent potentially new taxa. The cox1 analysis also suggested that HP88 is divided into two subgroups: the GPS11 group and the Oswego subgroup.