Successful Recovery of Nuclear Protein-Coding Genes from Small Insects in Museums Using Illumina Sequencing
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Wild Species 2010 the GENERAL STATUS of SPECIES in CANADA
Wild Species 2010 THE GENERAL STATUS OF SPECIES IN CANADA Canadian Endangered Species Conservation Council National General Status Working Group This report is a product from the collaboration of all provincial and territorial governments in Canada, and of the federal government. Canadian Endangered Species Conservation Council (CESCC). 2011. Wild Species 2010: The General Status of Species in Canada. National General Status Working Group: 302 pp. Available in French under title: Espèces sauvages 2010: La situation générale des espèces au Canada. ii Abstract Wild Species 2010 is the third report of the series after 2000 and 2005. The aim of the Wild Species series is to provide an overview on which species occur in Canada, in which provinces, territories or ocean regions they occur, and what is their status. Each species assessed in this report received a rank among the following categories: Extinct (0.2), Extirpated (0.1), At Risk (1), May Be At Risk (2), Sensitive (3), Secure (4), Undetermined (5), Not Assessed (6), Exotic (7) or Accidental (8). In the 2010 report, 11 950 species were assessed. Many taxonomic groups that were first assessed in the previous Wild Species reports were reassessed, such as vascular plants, freshwater mussels, odonates, butterflies, crayfishes, amphibians, reptiles, birds and mammals. Other taxonomic groups are assessed for the first time in the Wild Species 2010 report, namely lichens, mosses, spiders, predaceous diving beetles, ground beetles (including the reassessment of tiger beetles), lady beetles, bumblebees, black flies, horse flies, mosquitoes, and some selected macromoths. The overall results of this report show that the majority of Canada’s wild species are ranked Secure. -
A New Species of Bembidion Latrielle 1802 from the Ozarks, with a Review
A peer-reviewed open-access journal ZooKeys 147: 261–275 (2011)A new species of Bembidion Latrielle 1802 from the Ozarks... 261 doi: 10.3897/zookeys.147.1872 RESEARCH ARTICLE www.zookeys.org Launched to accelerate biodiversity research A new species of Bembidion Latrielle 1802 from the Ozarks, with a review of the North American species of subgenus Trichoplataphus Netolitzky 1914 (Coleoptera, Carabidae, Bembidiini) Drew A. Hildebrandt1,†, David R. Maddison2,‡ 1 710 Laney Road, Clinton, MS 39056 USA 2 Department of Zoology, Oregon State University, Corvallis, OR 97331, USA † urn:lsid:zoobank.org:author:038776CA-F70A-4744-96D6-B9B43FB56BB4 ‡ urn:lsid:zoobank.org:author:075A5E9B-5581-457D-8D2F-0B5834CDE04D Corresponding author: David R. Maddison ([email protected]) Academic editor: T. Erwin | Received 31 July 2011 | Accepted 25 August 2011 | Published 16 November 2011 urn:lsid:zoobank.org:pub:52038529-10EA-41A8-BE4F-6B495B610900 Citation: Hildebrandt DA, Maddison DR (2011) A new species of Bembidion Latrielle 1802 from the Ozarks, with a review of the North American species of subgenus Trichoplataphus Netolitzky 1914 (Coleoptera, Carabidae, Bembidiini). In: Erwin T (Ed) Proceedings of a symposium honoring the careers of Ross and Joyce Bell and their contributions to scientific work. Burlington, Vermont, 12–15 June 2010. ZooKeys 147: 261–275. doi: 10.3897/zookeys.147.1872 Abstract A new species of Bembidion (Trichoplataphus Netolitzky) from the Ozark Plateau of Missouri and Arkan- sas is described (Bembidion ozarkense Maddison and Hildebrandt). It is distinguishable from the closely related species, B. rolandi Fall, by characteristics of the male genitalia, and sequences of the genes cyto- chrome oxidase I and 28S ribosomal DNA. -
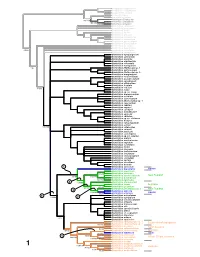
With Description of Bembidion Tahitiense, Sp. Nov. From
Asaphidion yukonense Asaphidion alaskanum Asaphidion curtum Lionepha erasa Lionepha osculans 99/100 Amerizus (Tiruka) sp. 100/100 Amerizus spectabilis 100/100 Amerizus wingatei Bembidion chalceum Bembidion properans Bembidion horni Bembidion hastii Bembidion planum Bembidion transversale Bembidion tetracolum Bembidion genei illigeri Bembidion geniculatum Bembidion cf. csikii Bembidion biguttatum 9 5/92 Bembidion wickhami Bembidion variegatum Bembidion ephippigerum Bembidion salinarium Bembidion assimile Bembidion nigrivestris Bembidion roosevelti Bembidion transparens 7 5/- Bembidion (Nothocys) sp. 1 Bembidion anthracinum Bembidion (Nothocys) sp. 2 Bembidion marginatum Bembidion fortestriatum Bembidion pseudocautum Bembidion canadianum Bembidion elizabethae Bembidion frontale 7 8/73 Bembidion siticum Bembidion lonae Bembidion sp. nr. lonae Bembidion melanopodum Bembidion scitulum Bembidion sexfoveatum Bembidion (Notholopha) sp. 1 Bembidion rugosellum Bembidion rawlinsi Bembidion rogersi Bembidion caoduroi Bembidion mandibulare Bembidion spinolai Bembidion chilense Bembidion sp. nr. chilense Bembidion hirtipes Bembidion rufoplagiatum Bembidion solieri 100/100 Bembidion cillenoides Bembidion calverti Bembidion posticale Bembidion convergens Bembidion sp. nr. ugartei Bembidion aratum Bembidion nubiculosum Bembidion rapidum Bembidion scintillans Bembidion flohri Bembidion idoneum Bembidion indistinctum Bembidion insulatum Bembidion obtusangulum Bembidion cordatum Bembidion varium Bembidion oberthueri Bembidion dorsale 5 Bembidion versutum -

Appendix O19749
Oikos o19749 Gerisch, M., Agostinelli, V., Henle, K. and Dziock, F. 2011. More species, but all do the same: contrasting effects of flood disturbance on ground beetle functional and species diversity. – Oikos 121: 508–515. Appendix A1 Tabelle1 Table A1. Full species list representing the standardized number of individuals per species for the study sites Steckby, Woerlitz, and Sandau. Density expresses the proportion of species standardized abundances to total abundance. Macropterous = winged, brachypterous = wingless, dimorphic = both forms can appear with a species. Body size is the average of maximum and minimum values found in the literature (for references see below). Wing Reproduction Body size Species names Steckby Woerlitz Sandau Density Morphology Season In mm Acupalpus dubius 0.032 0 0.016 0 macropterous spring 2.6 Acupalpus exiguus 1.838 1.019 0.71 0.005 macropterous spring 2.7 Acupalpus parvulus 0.081 0.038 0.032 0 macropterous spring 3.6 Agonum dolens 0.032 0.038 0.081 0 dimorph spring 8.8 Agonum duftschmidi 14.966 2.755 0.016 0.025 macropterous spring 8.2 Agonum emarginatum 116.659 4.472 25.194 0.208 macropterous spring 7.2 Agonum fuliginosum 0.097 0.038 0 0 dimorph spring 6.7 Agonum lugens 0.177 0 0.081 0 macropterous spring 9 Agonum marginatum 0.371 0.075 0.113 0.001 macropterous spring 9.2 Agonum micans 19.502 4.208 23.71 0.067 macropterous spring 6.6 Agonum muelleri 0 0.019 0 0 macropterous spring 8.2 Agonum piceum 0.468 0 0.016 0.001 macropterous spring 6.4 Agonum sexpunctatum 0.032 0 0.016 0 macropterous spring 8.2 Agonum -

Holocene Palaeoenvironmental Reconstruction Based on Fossil Beetle Faunas from the Altai-Xinjiang Region, China
Holocene palaeoenvironmental reconstruction based on fossil beetle faunas from the Altai-Xinjiang region, China Thesis submitted for the degree of Doctor of Philosophy at the University of London By Tianshu Zhang February 2018 Department of Geography, Royal Holloway, University of London Declaration of Authorship I Tianshu Zhang hereby declare that this thesis and the work presented in it is entirely my own. Where I have consulted the work of others, this is always clearly stated. Signed: Date: 25/02/2018 1 Abstract This project presents the results of the analysis of fossil beetle assemblages extracted from 71 samples from two peat profiles from the Halashazi Wetland in the southern Altai region of northwest China. The fossil assemblages allowed the reconstruction of local environments of the early (10,424 to 9500 cal. yr BP) and middle Holocene (6374 to 4378 cal. yr BP). In total, 54 Coleoptera taxa representing 44 genera and 14 families have been found, and 37 species have been identified, including a new species, Helophorus sinoglacialis. The majority of the fossil beetle species identified are today part of the Siberian fauna, and indicate cold steppe or tundra ecosystems. Based on the biogeographic affinities of the fossil faunas, it appears that the Altai Mountains served as dispersal corridor for cold-adapted (northern) beetle species during the Holocene. Quantified temperature estimates were made using the Mutual Climate Range (MCR) method. In addition, indicator beetle species (cold adapted species and bark beetles) have helped to identify both cold and warm intervals, and moisture conditions have been estimated on the basis of water associated species. -

A New Species of Bembidion LATREILLE from Nemrut Dağ
©Wiener Coleopterologenverein (WCV), download unter www.biologiezentrum.at Koleopterologische Rundschau 76 7–13 Wien, Juli 2006 A new species of Bembidion LATREILLE from Nemrut Da÷, Turkey (Coleoptera: Carabidae) L. TOLEDANO &K.RÉBL Abstract Bembidion (Ocydromus) nemrutdagi sp.n. from the Nemrut Da÷ (Turkey, AdÕyaman Prov.) is described. Its nearest relative, B. cordicolle DUVAL, 1851, is transferred to the subgen. Ocydromus CLAIRVILLE, 1806 s.l. (sensu KRYZHANOVSKIJ et al. 1995). The systematic relationships of some species groups are discussed. Key words: Coleoptera, Carabidae, Bembidiina, Bembidion, Ocydromus, Turkey, taxonomy. Introduction While studying Turkish Bembidion LATREILLE, 1802 we detected two specimens extremely similar to B. cordicolle DUVAL, 1851, from which they could be distinguished by the number of elytral pores. After comparison with the other species occurring in the area and after a thorough literature study we concluded that these specimens represent an undescribed species. Material and methods This paper is based on the study of 60 specimens belonging to the species dealt with herein and several hundreds of other specimens belonging to the subgen. Ocydromus CLAIRVILLE, 1806 sensu KRYZHANOVSKIJ et al. (1995). Collection Acronyms: CF Coll. Facchini, Piacenza CM Coll. Moret, Madrid CN Coll. Neri, S. Lorenzo in Noceto, Forlì CR Coll. Rébl, Nové Strašecí CT Coll. Toledano, Verona NMW Coll. Naturhistorisches Museum Wien The measurements, made with a Leica MZ12 stereobinocular microscope at 25 x (body) and 100 x (median lobes of aedeagi), are expressed in the text by the following abbreviations: el/ew elytral length / elytral width ratio ew/pw elytral width / pronotum width ratio pw/hw pronotum width / head width ratio pw/pl pronotum width / pronotum length ratio The body length has been measured from the front margin of the clypeus to the apex of the elytra, and the antennal length from the base of the antennomere 1 to the apex of antennomere 11. -

Amerizus (Tiruka) Gaoligongensis Sp. N. (Coleoptera: Carabidae): An
Research Article ISSN 2336-9744 (online) | ISSN 2337-0173 (print) The journal is available on line at www.ecol-mne.com http://zoobank.org/urn:lsid:zoobank.org:pub:4F9EE702-A3B4-47F9-83FF-ADD61BEE44AF Amerizus (Tiruka ) gaoligongensis sp. n. (Coleoptera: Carabidae): an endogean adapted representative of the genus BORISLAV GUÉORGUIEV National Museum of Natural History, 1 Blvd. Tzar Osvoboditel, 1000 Sofia, Bulgaria. E-mail: [email protected] Received 15 December 2014 │ Accepted 10 January 2015 │ Published online 12 January 2015. Abstract We describe and illustrate Amerizus (Tiruka) gaoligongensis sp. n. (type locality: cave Wu Shi Shan 1-2-3, near village Kongshuhe, Tengchong County, Yunnan, China). The species possesses a set of real troglomorphic characters, such as depigmentation of tegument, prolongation of the appendages, microphthalmy, roundness of both pronotum and elytra, and brachyptery. These special features, together with three corporal indexes and peculiarities in the structure of the aedeagus, enable a readily distinction of the new species. Otherwise, A. gaoligongensis seems related to species from the queinneci species-group. Key words : Coleoptera, Carabidae, Bembidiini, Amerizus , new species, China, Yunnan. Introduction The genus Amerizus Chaudoir, 1868 contains 50 species arranged in two subgenera and spread in Asia and North America (Sciaky and Toledano 2007; Bousquet 2012). One of the two subgenera, Tiruka Andrewes, 1935, includes 44 species, distributed from northwest India (state Jammu and Kashmir) to the central provinces of China. The species of this subgenus have been distributed into six groups of species (Sciaky and Toledano 2007; Toledano 2011). So far, twenty-two species of Tiruka were reported from China (Deuve 1998; Deuve 2002; Deuve 2004; Sciaky and Toledano 2007; Toledano 2011). -

Surveying for Terrestrial Arthropods (Insects and Relatives) Occurring Within the Kahului Airport Environs, Maui, Hawai‘I: Synthesis Report
Surveying for Terrestrial Arthropods (Insects and Relatives) Occurring within the Kahului Airport Environs, Maui, Hawai‘i: Synthesis Report Prepared by Francis G. Howarth, David J. Preston, and Richard Pyle Honolulu, Hawaii January 2012 Surveying for Terrestrial Arthropods (Insects and Relatives) Occurring within the Kahului Airport Environs, Maui, Hawai‘i: Synthesis Report Francis G. Howarth, David J. Preston, and Richard Pyle Hawaii Biological Survey Bishop Museum Honolulu, Hawai‘i 96817 USA Prepared for EKNA Services Inc. 615 Pi‘ikoi Street, Suite 300 Honolulu, Hawai‘i 96814 and State of Hawaii, Department of Transportation, Airports Division Bishop Museum Technical Report 58 Honolulu, Hawaii January 2012 Bishop Museum Press 1525 Bernice Street Honolulu, Hawai‘i Copyright 2012 Bishop Museum All Rights Reserved Printed in the United States of America ISSN 1085-455X Contribution No. 2012 001 to the Hawaii Biological Survey COVER Adult male Hawaiian long-horned wood-borer, Plagithmysus kahului, on its host plant Chenopodium oahuense. This species is endemic to lowland Maui and was discovered during the arthropod surveys. Photograph by Forest and Kim Starr, Makawao, Maui. Used with permission. Hawaii Biological Report on Monitoring Arthropods within Kahului Airport Environs, Synthesis TABLE OF CONTENTS Table of Contents …………….......................................................……………...........……………..…..….i. Executive Summary …….....................................................…………………...........……………..…..….1 Introduction ..................................................................………………………...........……………..…..….4 -

Ground Beetles in AB
J. Appl. Entomol. Identification of potential natural enemies of the pea leaf weevil, Sitona lineatus L. in western Canada M. A. Vankosky1,2,H.A.Ca´ rcamo2 & L. M. Dosdall1 1 Department of Agricultural, Food and Nutritional Science, University of Alberta, Edmonton, AB, Canada 2 Agriculture and Agri-Food Canada, Lethbridge Research Centre, Lethbridge, AB, Canada Keywords Abstract biological control, carabid ground beetles, conservation biological control, egg predation Sitona lineatus L., the pea leaf weevil, is an invasive pest of Pisum sativum L. (field pea) that has recently become established in Alberta, Canada. Correspondence Adults consume seedling foliage and larvae feed on Rhizobium root nod- M. A. Vankosky (corresponding author), ules thereby reducing nitrogen fixation; both life stages can reduce Department of Agricultural, Food and yield. Eggs and adults are vulnerable to predators and parasitoids. In Nutritional Science, 4-10 Agriculture-Forestry 2009, a series of experiments was undertaken to identify potential indig- Centre, University of Alberta, Edmonton, AB, Canada T6G 2P5. enous natural enemies of S. lineatus in southern Alberta. In three test E-mail: [email protected] arenas, eggs were exposed to starved ground beetles in no-choice tests for 48 h. Egg debris was observed when eggs were exposed to Bembidion Received: December 16, 2009; accepted: April quadrimaculatum L., Microlestes linearis (LeConte), Bembidion rupicola (Kir- 9, 2010. by), Bembidion timidum (LeConte), Poecilus scitulus LeConte, and staphyli- nid beetles. Of the two most abundant carabid species, the smaller doi: 10.1111/j.1439-0418.2010.01542.x B. quadrimaculatum consistently removed significantly more eggs (94.6%) than the larger Pterostichus melanarius (17.4%). -

The Works of Blackburn and Sharp Established the Greater Part
235 The Carabid Tribes Harpalini, Lebiini and Bembidiini in Hawaii (Coleoptera) By EVERARD B. BRITTON Department of Entomology, British Museum (Natural History) (Presented by B. C. Zimmerman at the meeting of December 8, 1947) The works of Blackburn and Sharp established the greater part of the species of Hawaiian Carabidae, but identification has re mained an impossibility without reference to the original series. As the types of the species described by both Blackburn and Sharp are in the British Museum, I have undertaken a revision of the carabid fauna of the Islands. The Nomiini have been the object of a special report recently published by Bishop Museum (Bishop Mus. Occ. Papers, 19 (4): 107-166, figs. 1-17, 1948). Work is now m progress on the remaining tribe, the Anchomenini. I wish to express my thanks to Mr. E. C. Zimmerman for colla boration and advice, and for his care in the preparation of this paper for the press. It was at his suggestion that this work was begun, and in 1938 he turned over to me his manuscript revision of the Hawaiian Carabidae which he was unable to complete because of the lack of types in Honolulu. The extreme isolation of the Hawaiian Islands is clearly reflected in the carabid fauna. Of the fifty or more tribes of Carabidae which are recognized, only five are represented in the Islands, and only three have existed there long enough to permit the evolution of endemic species. The tribes with endemic species are the Bembi diini, Nomiini and Anchomenini. The Nomiini and Anchomenini, which are not dealt with here, each comprise a single genus with a large number of species, alt of which are endemic. -

The Effects of Spatial and Temporal Heterogeneity on the Population Dynamics of Four Animal Species in a Danish Landscape
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Valery Forbes Publications Papers in the Biological Sciences 6-23-2009 The effects of spatial and temporal heterogeneity on the population dynamics of four animal species in a Danish landscape Richard M. Sibly University of Reading, [email protected] Jacob Nabe-Nielsen University of Aarhus, Roskilde, Denmark, [email protected] Mads C. Forchhammer University of Aarhus, Roskilde, Denmark, [email protected] Valery E. Forbes University of Nebraska-Lincoln, [email protected] Christopher J. Topping University of Aarhus, Rønde, Denmark, [email protected] Follow this and additional works at: https://digitalcommons.unl.edu/biosciforbes Part of the Pharmacology, Toxicology and Environmental Health Commons Sibly, Richard M.; Nabe-Nielsen, Jacob; Forchhammer, Mads C.; Forbes, Valery E.; and Topping, Christopher J., "The effects of spatial and temporal heterogeneity on the population dynamics of four animal species in a Danish landscape" (2009). Valery Forbes Publications. 12. https://digitalcommons.unl.edu/biosciforbes/12 This Article is brought to you for free and open access by the Papers in the Biological Sciences at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Valery Forbes Publications by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. BMC Ecology BioMed Central Research article Open Access The effects of spatial and temporal heterogeneity on the population dynamics of four animal species in a Danish landscape -

Fauna Europaea: Coleoptera 2 (Excl. Series Elateriformia, Scarabaeiformia, Staphyliniformia and Superfamily Curculionoidea)
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Digital.CSIC Biodiversity Data Journal 3: e4750 doi: 10.3897/BDJ.3.e4750 Data Paper Fauna Europaea: Coleoptera 2 (excl. series Elateriformia, Scarabaeiformia, Staphyliniformia and superfamily Curculionoidea) Paolo Audisio‡, Miguel-Angel Alonso Zarazaga§, Adam Slipinski|, Anders Nilsson¶#, Josef Jelínek , Augusto Vigna Taglianti‡, Federica Turco ¤, Carlos Otero«, Claudio Canepari», David Kral ˄, Gianfranco Liberti˅, Gianfranco Sama¦, Gianluca Nardi ˀ, Ivan Löblˁ, Jan Horak ₵, Jiri Kolibacℓ, Jirí Háva ₰, Maciej Sapiejewski†,₱, Manfred Jäch ₳, Marco Alberto Bologna₴, Maurizio Biondi ₣, Nikolai B. Nikitsky₮, Paolo Mazzoldi₦, Petr Zahradnik ₭, Piotr Wegrzynowicz₱, Robert Constantin₲, Roland Gerstmeier‽, Rustem Zhantiev₮, Simone Fattorini₩, Wioletta Tomaszewska₱, Wolfgang H. Rücker₸, Xavier Vazquez- Albalate‡‡, Fabio Cassola §§, Fernando Angelini||, Colin Johnson ¶¶, Wolfgang Schawaller##, Renato Regalin¤¤, Cosimo Baviera««, Saverio Rocchi »», Fabio Cianferoni»»,˄˄, Ron Beenen ˅˅, Michael Schmitt ¦¦, David Sassi ˀˀ, Horst Kippenbergˁˁ, Marcello Franco Zampetti₩, Marco Trizzino ₵₵, Stefano Chiari‡, Giuseppe Maria Carpanetoℓℓ, Simone Sabatelli‡, Yde de Jong ₰₰,₱₱ ‡ Sapienza Rome University, Department of Biology and Biotechnologies 'C. Darwin', Rome, Italy § Museo Nacional de Ciencias Naturales, Madrid, Spain | CSIRO Entomology, Canberra, Australia ¶ Umea University, Umea, Sweden # National Museum Prague, Prague, Czech Republic ¤ Queensland Museum, Brisbane,