Blueprint Genetics Microcephaly and Pontocerebellar Hypoplasia Panel
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
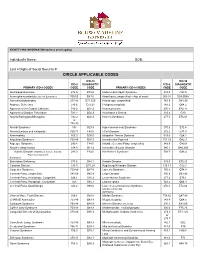
Circle Applicable Codes
IDENTIFYING INFORMATION (please print legibly) Individual’s Name: DOB: Last 4 Digits of Social Security #: CIRCLE APPLICABLE CODES ICD-10 ICD-10 ICD-9 DIAGNOSTIC ICD-9 DIAGNOSTIC PRIMARY ICD-9 CODES CODE CODE PRIMARY ICD-9 CODES CODE CODE Abetalipoproteinemia 272.5 E78.6 Hallervorden-Spatz Syndrome 333.0 G23.0 Acrocephalosyndactyly (Apert’s Syndrome) 755.55 Q87.0 Head Injury, unspecified – Age of onset: 959.01 S09.90XA Adrenaleukodystrophy 277.86 E71.529 Hemiplegia, unspecified 342.9 G81.90 Arginase Deficiency 270.6 E72.21 Holoprosencephaly 742.2 Q04.2 Agenesis of the Corpus Callosum 742.2 Q04.3 Homocystinuria 270.4 E72.11 Agenesis of Septum Pellucidum 742.2 Q04.3 Huntington’s Chorea 333.4 G10 Argyria/Pachygyria/Microgyria 742.2 Q04.3 Hurler’s Syndrome 277.5 E76.01 or 758.33 Aicardi Syndrome 333 G23.8 Hyperammonemia Syndrome 270.6 E72.4 Alcohol Embryo and Fetopathy 760.71 F84.5 I-Cell Disease 272.2 E77.0 Anencephaly 655.0 Q00.0 Idiopathic Torsion Dystonia 333.6 G24.1 Angelman Syndrome 759.89 Q93.5 Incontinentia Pigmenti 757.33 Q82.3 Asperger Syndrome 299.8 F84.5 Infantile Cerebral Palsy, unspecified 343.9 G80.9 Ataxia-Telangiectasia 334.8 G11.3 Intractable Seizure Disorder 345.1 G40.309 Autistic Disorder (Childhood Autism, Infantile 299.0 F84.0 Klinefelter’s Syndrome 758.7 Q98.4 Psychosis, Kanner’s Syndrome) Biotinidase Deficiency 277.6 D84.1 Krabbe Disease 333.0 E75.23 Canavan Disease 330.0 E75.29 Kugelberg-Welander Disease 335.11 G12.1 Carpenter Syndrome 759.89 Q87.0 Larsen’s Syndrome 755.8 Q74.8 Cerebral Palsy, unspecified 343.69 G80.9 -

TARGETED GENE PANELS and REFERENCE SEQUENCE (Refseq) TRANSCRIPTS by PHENOTYPE
ROYAL DEVON & EXETER NHS FOUNDATION TRUST Department of Molecular Genetics TARGETED GENE PANELS AND REFERENCE SEQUENCE (RefSeq) TRANSCRIPTS BY PHENOTYPE Table of Contents (Click to select) Alagille Syndrome 2 Chondrodysplasia punctata 2 Combined Pituitary Hormone Deficiency 2 Congenital Generalised Lipodystrophy 2 Congenital Hypothyroidism 2 Early-onset Diabetes and Autoimmunity 3 Endocrine Neoplasia Syndromes 3 Familial Glucocorticoid Deficiency 3 Familial Hyperparathyroidism 3 Familial Hypocalciuric Hypercalcaemia 3 Familial Hyperparathyroidism/hypercalcaemia 4 Familial Hypoparathyroidism 4 Familial Partial Lipodystrophy 4 Familial Porencephaly and HANAC syndrome 4 Familial Tumoral Calcinosis 4 Feingold syndrome 4 Gastrointestinal atresia 5 Generalised Arterial Calcification in Infancy 5 Holoprosencephaly 5 Hyperinsulinism 5 Hypophosphatemic Rickets 6 Isolated Growth Hormone Deficiency 6 Kabuki syndrome 6 Kallmann syndrome 6 Mandibulofacial Dysostosis with Microcephaly 6 Moebius syndrome 6 Monogenic Diabetes of the Young (MODY) 7 Multiple Exostosis 7 Neonatal Diabetes 8 Phaeochromocytoma/Paraganglioma 9 Pontocerebellar Hypoplasia 9 Primary pigmented nodular adrenocortical disease 9 Pseudohypoaldosteronism 9 Spondylocostal Dysostosis 9 Visceral heterotaxy 10 Page 1 of 10 ROYAL DEVON & EXETER NHS FOUNDATION TRUST Department of Molecular Genetics Alagille Syndrome Transcript(s) JAG1 NM_000214 NOTCH2 NM_024408 Chondrodysplasia punctata Transcript(s) AGPS NM_003659 ARSE NM_000047 EBP NM_006579 GNPAT NM_014236 PEX7 NM_000288 Combined Pituitary -

WES Gene Package Multiple Congenital Anomalie.Xlsx
Whole Exome Sequencing Gene package Multiple congenital anomalie, version 5, 1‐2‐2018 Technical information DNA was enriched using Agilent SureSelect Clinical Research Exome V2 capture and paired‐end sequenced on the Illumina platform (outsourced). The aim is to obtain 8.1 Giga base pairs per exome with a mapped fraction of 0.99. The average coverage of the exome is ~50x. Duplicate reads are excluded. Data are demultiplexed with bcl2fastq Conversion Software from Illumina. Reads are mapped to the genome using the BWA‐MEM algorithm (reference: http://bio‐bwa.sourceforge.net/). Variant detection is performed by the Genome Analysis Toolkit HaplotypeCaller (reference: http://www.broadinstitute.org/gatk/). The detected variants are filtered and annotated with Cartagenia software and classified with Alamut Visual. It is not excluded that pathogenic mutations are being missed using this technology. At this moment, there is not enough information about the sensitivity of this technique with respect to the detection of deletions and duplications of more than 5 nucleotides and of somatic mosaic mutations (all types of sequence changes). HGNC approved Phenotype description including OMIM phenotype ID(s) OMIM median depth % covered % covered % covered gene symbol gene ID >10x >20x >30x A4GALT [Blood group, P1Pk system, P(2) phenotype], 111400 607922 101 100 100 99 [Blood group, P1Pk system, p phenotype], 111400 NOR polyagglutination syndrome, 111400 AAAS Achalasia‐addisonianism‐alacrimia syndrome, 231550 605378 73 100 100 100 AAGAB Keratoderma, palmoplantar, -

MECHANISMS in ENDOCRINOLOGY: Novel Genetic Causes of Short Stature
J M Wit and others Genetics of short stature 174:4 R145–R173 Review MECHANISMS IN ENDOCRINOLOGY Novel genetic causes of short stature 1 1 2 2 Jan M Wit , Wilma Oostdijk , Monique Losekoot , Hermine A van Duyvenvoorde , Correspondence Claudia A L Ruivenkamp2 and Sarina G Kant2 should be addressed to J M Wit Departments of 1Paediatrics and 2Clinical Genetics, Leiden University Medical Center, PO Box 9600, 2300 RC Leiden, Email The Netherlands [email protected] Abstract The fast technological development, particularly single nucleotide polymorphism array, array-comparative genomic hybridization, and whole exome sequencing, has led to the discovery of many novel genetic causes of growth failure. In this review we discuss a selection of these, according to a diagnostic classification centred on the epiphyseal growth plate. We successively discuss disorders in hormone signalling, paracrine factors, matrix molecules, intracellular pathways, and fundamental cellular processes, followed by chromosomal aberrations including copy number variants (CNVs) and imprinting disorders associated with short stature. Many novel causes of GH deficiency (GHD) as part of combined pituitary hormone deficiency have been uncovered. The most frequent genetic causes of isolated GHD are GH1 and GHRHR defects, but several novel causes have recently been found, such as GHSR, RNPC3, and IFT172 mutations. Besides well-defined causes of GH insensitivity (GHR, STAT5B, IGFALS, IGF1 defects), disorders of NFkB signalling, STAT3 and IGF2 have recently been discovered. Heterozygous IGF1R defects are a relatively frequent cause of prenatal and postnatal growth retardation. TRHA mutations cause a syndromic form of short stature with elevated T3/T4 ratio. Disorders of signalling of various paracrine factors (FGFs, BMPs, WNTs, PTHrP/IHH, and CNP/NPR2) or genetic defects affecting cartilage extracellular matrix usually cause disproportionate short stature. -

(12) Patent Application Publication (10) Pub. No.: US 2016/0281166 A1 BHATTACHARJEE Et Al
US 20160281 166A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2016/0281166 A1 BHATTACHARJEE et al. (43) Pub. Date: Sep. 29, 2016 (54) METHODS AND SYSTEMIS FOR SCREENING Publication Classification DISEASES IN SUBJECTS (51) Int. Cl. (71) Applicant: PARABASE GENOMICS, INC., CI2O I/68 (2006.01) Boston, MA (US) C40B 30/02 (2006.01) (72) Inventors: Arindam BHATTACHARJEE, G06F 9/22 (2006.01) Andover, MA (US); Tanya (52) U.S. Cl. SOKOLSKY, Cambridge, MA (US); CPC ............. CI2O 1/6883 (2013.01); G06F 19/22 Edwin NAYLOR, Mt. Pleasant, SC (2013.01); C40B 30/02 (2013.01); C12O (US); Richard B. PARAD, Newton, 2600/156 (2013.01); C12O 2600/158 MA (US); Evan MAUCELI, (2013.01) Roslindale, MA (US) (21) Appl. No.: 15/078,579 (57) ABSTRACT (22) Filed: Mar. 23, 2016 Related U.S. Application Data The present disclosure provides systems, devices, and meth (60) Provisional application No. 62/136,836, filed on Mar. ods for a fast-turnaround, minimally invasive, and/or cost 23, 2015, provisional application No. 62/137,745, effective assay for Screening diseases, such as genetic dis filed on Mar. 24, 2015. orders and/or pathogens, in Subjects. Patent Application Publication Sep. 29, 2016 Sheet 1 of 23 US 2016/0281166 A1 SSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS S{}}\\93? sau36 Patent Application Publication Sep. 29, 2016 Sheet 2 of 23 US 2016/0281166 A1 &**** ? ???zzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz??º & %&&zzzzzzzzzzzzzzzzzzzzzzz &Sssssssssssssssssssssssssssssssssssssssssssssssssssssssss & s s sS ------------------------------ Patent Application Publication Sep. 29, 2016 Sheet 3 of 23 US 2016/0281166 A1 23 25 20 FG, 2. Patent Application Publication Sep. 29, 2016 Sheet 4 of 23 US 2016/0281166 A1 : S Patent Application Publication Sep. -

Genetic Short Stature ⁎ Michelle Grunauera, Alexander A.L
Growth Hormone & IGF Research xxx (xxxx) xxx–xxx Contents lists available at ScienceDirect Growth Hormone & IGF Research journal homepage: www.elsevier.com/locate/ghir Genetic short stature ⁎ Michelle Grunauera, Alexander A.L. Jorgeb, a Pediatric Intensive Care Unit, Hospital de los Valles, Escuela de Medicina, Universidad San Francisco de Quito (USFQ), Quito, Ecuador b Unidade de Endocrinologia Genetica (LIM25), Hospital das Clinicas da Faculdade de Medicina da Universidade de São Paulo (FMUSP), Sao Paulo, Brazil ARTICLE INFO ABSTRACT Keywords: Adult height and growth patterns are largely genetically programmed. Studies in twins have indicated that the Genetic heritability of height is high (> 80%), suggesting that genetic variation is the main determinant of stature. Short stature Height exhibits a normal (Gaussian) distribution according to sex, age, and ancestry. Short stature is usually Skeletal dysplasia defined as a height which is 2 standard deviations (S.D.) less than the mean height of a specific population. This Syndrome definition includes 2.3% of the population and usually includes healthy individuals. In this group of short stature Adult height non-syndromic conditions, the genetic influence occurs polygenically or oligogenically. As a rule, each common genetic variant accounts for a small effect (1 mm) on individual height variation. Recently, several studies de- monstrated that some rare variants can cause greater effect on height, without causing a syndromic condition. In more extreme cases, height SDS below 2.5 or 3 (which would comprise approximately 0.6 and 0.1% of the population, respectively) is frequently associated with syndromic conditions and are usually caused by a monogenic defect. -

Blueprint Genetics Comprehensive Growth Disorders / Skeletal
Comprehensive Growth Disorders / Skeletal Dysplasias and Disorders Panel Test code: MA4301 Is a 374 gene panel that includes assessment of non-coding variants. This panel covers the majority of the genes listed in the Nosology 2015 (PMID: 26394607) and all genes in our Malformation category that cause growth retardation, short stature or skeletal dysplasia and is therefore a powerful diagnostic tool. It is ideal for patients suspected to have a syndromic or an isolated growth disorder or a skeletal dysplasia. About Comprehensive Growth Disorders / Skeletal Dysplasias and Disorders This panel covers a broad spectrum of diseases associated with growth retardation, short stature or skeletal dysplasia. Many of these conditions have overlapping features which can make clinical diagnosis a challenge. Genetic diagnostics is therefore the most efficient way to subtype the diseases and enable individualized treatment and management decisions. Moreover, detection of causative mutations establishes the mode of inheritance in the family which is essential for informed genetic counseling. For additional information regarding the conditions tested on this panel, please refer to the National Organization for Rare Disorders and / or GeneReviews. Availability 4 weeks Gene Set Description Genes in the Comprehensive Growth Disorders / Skeletal Dysplasias and Disorders Panel and their clinical significance Gene Associated phenotypes Inheritance ClinVar HGMD ACAN# Spondyloepimetaphyseal dysplasia, aggrecan type, AD/AR 20 56 Spondyloepiphyseal dysplasia, Kimberley -

Blueprint Genetics Comprehensive Skeletal Dysplasias and Disorders
Comprehensive Skeletal Dysplasias and Disorders Panel Test code: MA3301 Is a 251 gene panel that includes assessment of non-coding variants. Is ideal for patients with a clinical suspicion of disorders involving the skeletal system. About Comprehensive Skeletal Dysplasias and Disorders This panel covers a broad spectrum of skeletal disorders including common and rare skeletal dysplasias (eg. achondroplasia, COL2A1 related dysplasias, diastrophic dysplasia, various types of spondylo-metaphyseal dysplasias), various ciliopathies with skeletal involvement (eg. short rib-polydactylies, asphyxiating thoracic dysplasia dysplasias and Ellis-van Creveld syndrome), various subtypes of osteogenesis imperfecta, campomelic dysplasia, slender bone dysplasias, dysplasias with multiple joint dislocations, chondrodysplasia punctata group of disorders, neonatal osteosclerotic dysplasias, osteopetrosis and related disorders, abnormal mineralization group of disorders (eg hypopohosphatasia), osteolysis group of disorders, disorders with disorganized development of skeletal components, overgrowth syndromes with skeletal involvement, craniosynostosis syndromes, dysostoses with predominant craniofacial involvement, dysostoses with predominant vertebral involvement, patellar dysostoses, brachydactylies, some disorders with limb hypoplasia-reduction defects, ectrodactyly with and without other manifestations, polydactyly-syndactyly-triphalangism group of disorders, and disorders with defects in joint formation and synostoses. Availability 4 weeks Gene Set Description -

Review of Genetics and Epidemiology. Charles Shaw
JMG Online First, published on November 21, 2005 as 10.1136/jmg.2005.038158 J Med Genet: first published as 10.1136/jmg.2005.038158 on 18 November 2005. Downloaded from Oesophageal atresia, tracheo-oesophageal fistula and the VACTERL association: review of genetics and epidemiology. Charles Shaw-Smith Department of Medical Genetics, Addenbrooke’s Hospital, Cambridge CB2 2QQ, Address for correspondence: Charles Shaw-Smith Department of Medical Genetics, Box 134, Addenbrooke’s Hospital, Hills Road, Cambridge CB2 2QQ UK Telephone +44(0)1223 216446 Fax +44(0)1223 217054 Email [email protected] http://jmg.bmj.com/ Running title: Genetics of oesophageal atresia on September 26, 2021 by guest. Protected copyright. Copyright Article author (or their employer) 2005. Produced by BMJ Publishing Group Ltd under licence. J Med Genet: first published as 10.1136/jmg.2005.038158 on 18 November 2005. Downloaded from Abstract Oesophageal atresia and/or tracheo-oesophageal fistula are common malformations occurring in approximately 1 in 3500 births. In around half of cases (syndromic oesophageal atresia), there are other associated anomalies, with cardiac malformations being the most common. These may occur as part of the VACTERL association (OMIM 192350). In the remainder of cases, oesophageal atresia/tracheo-oesophageal fistula occur in isolation (non-syndromic oesophageal atresia). Data from twin and family studies suggest that, in the majority of cases, genetic factors do not play a major role, and yet there are well-defined instances of this malformation where genetic factors clearly are important. This is highlighted by the recent identification of no fewer than three separate genes with a role in the aetiology of oesophageal atresia. -

Supplementary Information
Supplementary Information Structural Capacitance in Protein Evolution and Human Diseases Chen Li, Liah V T Clark, Rory Zhang, Benjamin T Porebski, Julia M. McCoey, Natalie A. Borg, Geoffrey I. Webb, Itamar Kass, Malcolm Buckle, Jiangning Song, Adrian Woolfson, and Ashley M. Buckle Supplementary tables Table S1. Disorder prediction using the human disease and polymorphisms dataseta OR DR OO OD DD DO mutations mutations 24,758 650 2,741 513 Disease 25,408 3,254 97.44% 2.56% 84.23% 15.77% 26,559 809 11,135 1,218 Non-disease 27,368 12,353 97.04% 2.96% 90.14% 9.86% ahttp://www.uniprot.org/docs/humsavar [1] (see Materials and Methdos). The numbers listed are the ones of unique mutations. ‘Unclassifiied’ mutations, according to the UniProt, were not counted. O = predicted as ordered; OR = Ordered regions D = predicted as disordered; DR = Disordered regions 1 Table S2. Mutations in long disordered regions (LDRs) of human proteins predicted to produce a DO transitiona Average # disorder # disorder # disorder # order UniProt/dbSNP Protein Mutation Disease length of predictors predictors predictorsb predictorsc LDRd in D2P2e for LDRf UHRF1-binding protein 1- A0JNW5/rs7296162 like S1147L - 4 2^ 101 6 3 A4D1E1/rs801841 Zinc finger protein 804B V1195I - 3* 2^ 37 6 1 A6NJV1/rs2272466 UPF0573 protein C2orf70 Q177L - 2* 4 34 3 1 Golgin subfamily A member A7E2F4/rs347880 8A K480N - 2* 2^ 91 N/A 2 Axonemal dynein light O14645/rs11749 intermediate polypeptide 1 A65V - 3* 3 43 N/A 2 Centrosomal protein of 290 O15078/rs374852145 kDa R2210C - 2 3 123 5 1 Fanconi -

Soonerstart Automatic Qualifying Syndromes and Conditions
SoonerStart Automatic Qualifying Syndromes and Conditions - Appendix O Abetalipoproteinemia Acanthocytosis (see Abetalipoproteinemia) Accutane, Fetal Effects of (see Fetal Retinoid Syndrome) Acidemia, 2-Oxoglutaric Acidemia, Glutaric I Acidemia, Isovaleric Acidemia, Methylmalonic Acidemia, Propionic Aciduria, 3-Methylglutaconic Type II Aciduria, Argininosuccinic Acoustic-Cervico-Oculo Syndrome (see Cervico-Oculo-Acoustic Syndrome) Acrocephalopolysyndactyly Type II Acrocephalosyndactyly Type I Acrodysostosis Acrofacial Dysostosis, Nager Type Adams-Oliver Syndrome (see Limb and Scalp Defects, Adams-Oliver Type) Adrenoleukodystrophy, Neonatal (see Cerebro-Hepato-Renal Syndrome) Aglossia Congenita (see Hypoglossia-Hypodactylia) Aicardi Syndrome AIDS Infection (see Fetal Acquired Immune Deficiency Syndrome) Alaninuria (see Pyruvate Dehydrogenase Deficiency) Albers-Schonberg Disease (see Osteopetrosis, Malignant Recessive) Albinism, Ocular (includes Autosomal Recessive Type) Albinism, Oculocutaneous, Brown Type (Type IV) Albinism, Oculocutaneous, Tyrosinase Negative (Type IA) Albinism, Oculocutaneous, Tyrosinase Positive (Type II) Albinism, Oculocutaneous, Yellow Mutant (Type IB) Albinism-Black Locks-Deafness Albright Hereditary Osteodystrophy (see Parathyroid Hormone Resistance) Alexander Disease Alopecia - Mental Retardation Alpers Disease Alpha 1,4 - Glucosidase Deficiency (see Glycogenosis, Type IIA) Alpha-L-Fucosidase Deficiency (see Fucosidosis) Alport Syndrome (see Nephritis-Deafness, Hereditary Type) Amaurosis (see Blindness) Amaurosis -

VACTERL/VATER Association Benjamin D Solomon
Solomon Orphanet Journal of Rare Diseases 2011, 6:56 http://www.ojrd.com/content/6/1/56 REVIEW Open Access VACTERL/VATER Association Benjamin D Solomon Abstract VACTERL/VATER association is typically defined by the presence of at least three of the following congenital malformations: vertebral defects, anal atresia, cardiac defects, tracheo-esophageal fistula, renal anomalies, and limb abnormalities. In addition to these core component features, patients may also have other congenital anomalies. Although diagnostic criteria vary, the incidence is estimated at approximately 1 in 10,000 to 1 in 40,000 live-born infants. The condition is ascertained clinically by the presence of the above-mentioned malformations; importantly, there should be no clinical or laboratory-based evidence for the presence of one of the many similar conditions, as the differential diagnosis is relatively large. This differential diagnosis includes (but is not limited to) Baller-Gerold syndrome, CHARGE syndrome, Currarino syndrome, deletion 22q11.2 syndrome, Fanconi anemia, Feingold syndrome, Fryns syndrome, MURCS association, oculo-auriculo-vertebral syndrome, Opitz G/BBB syndrome, Pallister- Hall syndrome, Townes-Brocks syndrome, and VACTERL with hydrocephalus. Though there are hints regarding causation, the aetiology has been identified only in a small fraction of patients to date, likely due to factors such as a high degree of clinical and causal heterogeneity, the largely sporadic nature of the disorder, and the presence of many similar conditions. New genetic research methods offer promise that the causes of VACTERL association will be better defined in the relatively near future. Antenatal diagnosis can be challenging, as certain component features can be difficult to ascertain prior to birth.