Supplementary Information
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Chuannaky2016phd.Pdf (13.10Mb)
! " " ZINC%HOMEOSTASIS!IN#HEALTH,!EXERCISE!AND$ CHRONIC'DISEASES! " " " " " " Anna Kit Yung Chu BAppSc (Ex&SpSc) / BSc (Nutr) (Hons I) The University of Sydney, 2012 A thesis submitted in fulfilment of the requirements for the degree of Doctor of Philosophy (PhD) Department of Human Nutrition Division of Sciences University of Otago 2016 i" " """""" """""! " Table&of&Contents& & Abstract&...................................................................................................................................&iv" Acknowledgments&...............................................................................................................&vi" List&of&abbreviations&........................................................................................................&viii" List&of&tables&............................................................................................................................&x" List&of&figures&.......................................................................................................................&xii" Publications&arising&from&this&thesis&............................................................................&xv" List&of&other&publications&arising&from&this&thesis&.................................................&xvi" Chapter&1&&&Introduction&and&Literature&Review&.............................................................&1" Introduction&............................................................................................................................&2" Whole&body&zinc&homeostasis&...........................................................................................&5" -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Loss of the Dermis Zinc Transporter ZIP13 Promotes the Mildness Of
www.nature.com/scientificreports OPEN Loss of the dermis zinc transporter ZIP13 promotes the mildness of fbrosarcoma by inhibiting autophagy Mi-Gi Lee1,8, Min-Ah Choi2,8, Sehyun Chae3,8, Mi-Ae Kang4, Hantae Jo4, Jin-myoung Baek4, Kyu-Ree In4, Hyein Park4, Hyojin Heo4, Dongmin Jang5, Sofa Brito4, Sung Tae Kim6, Dae-Ok Kim 1,7, Jong-Soo Lee4, Jae-Ryong Kim2* & Bum-Ho Bin 4* Fibrosarcoma is a skin tumor that is frequently observed in humans, dogs, and cats. Despite unsightly appearance, studies on fbrosarcoma have not signifcantly progressed, due to a relatively mild tumor severity and a lower incidence than that of other epithelial tumors. Here, we focused on the role of a recently-found dermis zinc transporter, ZIP13, in fbrosarcoma progression. We generated two transformed cell lines from wild-type and ZIP13-KO mice-derived dermal fbroblasts by stably expressing the Simian Virus (SV) 40-T antigen. The ZIP13−/− cell line exhibited an impairment in autophagy, followed by hypersensitivity to nutrient defciency. The autophagy impairment in the ZIP13−/− cell line was due to the low expression of LC3 gene and protein, and was restored by the DNA demethylating agent, 5-aza-2’-deoxycytidine (5-aza) treatment. Moreover, the DNA methyltransferase activity was signifcantly increased in the ZIP13−/− cell line, indicating the disturbance of epigenetic regulations. Autophagy inhibitors efectively inhibited the growth of fbrosarcoma with relatively minor damages to normal cells in xenograft assay. Our data show that proper control over autophagy and zinc homeostasis could allow for the development of a new therapeutic strategy to treat fbrosarcoma. -
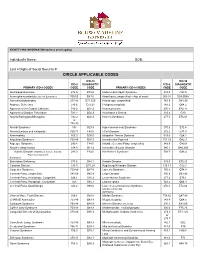
Circle Applicable Codes
IDENTIFYING INFORMATION (please print legibly) Individual’s Name: DOB: Last 4 Digits of Social Security #: CIRCLE APPLICABLE CODES ICD-10 ICD-10 ICD-9 DIAGNOSTIC ICD-9 DIAGNOSTIC PRIMARY ICD-9 CODES CODE CODE PRIMARY ICD-9 CODES CODE CODE Abetalipoproteinemia 272.5 E78.6 Hallervorden-Spatz Syndrome 333.0 G23.0 Acrocephalosyndactyly (Apert’s Syndrome) 755.55 Q87.0 Head Injury, unspecified – Age of onset: 959.01 S09.90XA Adrenaleukodystrophy 277.86 E71.529 Hemiplegia, unspecified 342.9 G81.90 Arginase Deficiency 270.6 E72.21 Holoprosencephaly 742.2 Q04.2 Agenesis of the Corpus Callosum 742.2 Q04.3 Homocystinuria 270.4 E72.11 Agenesis of Septum Pellucidum 742.2 Q04.3 Huntington’s Chorea 333.4 G10 Argyria/Pachygyria/Microgyria 742.2 Q04.3 Hurler’s Syndrome 277.5 E76.01 or 758.33 Aicardi Syndrome 333 G23.8 Hyperammonemia Syndrome 270.6 E72.4 Alcohol Embryo and Fetopathy 760.71 F84.5 I-Cell Disease 272.2 E77.0 Anencephaly 655.0 Q00.0 Idiopathic Torsion Dystonia 333.6 G24.1 Angelman Syndrome 759.89 Q93.5 Incontinentia Pigmenti 757.33 Q82.3 Asperger Syndrome 299.8 F84.5 Infantile Cerebral Palsy, unspecified 343.9 G80.9 Ataxia-Telangiectasia 334.8 G11.3 Intractable Seizure Disorder 345.1 G40.309 Autistic Disorder (Childhood Autism, Infantile 299.0 F84.0 Klinefelter’s Syndrome 758.7 Q98.4 Psychosis, Kanner’s Syndrome) Biotinidase Deficiency 277.6 D84.1 Krabbe Disease 333.0 E75.23 Canavan Disease 330.0 E75.29 Kugelberg-Welander Disease 335.11 G12.1 Carpenter Syndrome 759.89 Q87.0 Larsen’s Syndrome 755.8 Q74.8 Cerebral Palsy, unspecified 343.69 G80.9 -

Restoration of Epithelial Sodium Channel Function by Synthetic Peptides in Pseudohypoaldosteronism Type 1B Mutants
ORIGINAL RESEARCH published: 24 February 2017 doi: 10.3389/fphar.2017.00085 Restoration of Epithelial Sodium Channel Function by Synthetic Peptides in Pseudohypoaldosteronism Type 1B Mutants Anita Willam 1*, Mohammed Aufy 1, Susan Tzotzos 2, Heinrich Evanzin 1, Sabine Chytracek 1, Sabrina Geppert 1, Bernhard Fischer 2, Hendrik Fischer 2, Helmut Pietschmann 2, Istvan Czikora 3, Rudolf Lucas 3, Rosa Lemmens-Gruber 1 and Waheed Shabbir 1, 2 1 Department of Pharmacology and Toxicology, University of Vienna, Vienna, Austria, 2 APEPTICO GmbH, Vienna, Austria, 3 Vascular Biology Center, Medical College of Georgia, Augusta University, Augusta, GA, USA Edited by: The synthetically produced cyclic peptides solnatide (a.k.a. TIP or AP301) and its Gildas Loussouarn, congener AP318, whose molecular structures mimic the lectin-like domain of human University of Nantes, France tumor necrosis factor (TNF), have been shown to activate the epithelial sodium Reviewed by: channel (ENaC) in various cell- and animal-based studies. Loss-of-ENaC-function Stephan Kellenberger, University of Lausanne, Switzerland leads to a rare, life-threatening, salt-wasting syndrome, pseudohypoaldosteronism type Yoshinori Marunaka, 1B (PHA1B), which presents with failure to thrive, dehydration, low blood pressure, Kyoto Prefectural University of Medicine, Japan anorexia and vomiting; hyperkalemia, hyponatremia and metabolic acidosis suggest *Correspondence: hypoaldosteronism, but plasma aldosterone and renin activity are high. The aim of Anita Willam the present study was to investigate whether the ENaC-activating effect of solnatide [email protected] and AP318 could rescue loss-of-function phenotype of ENaC carrying mutations at + Specialty section: conserved amino acid positions observed to cause PHA1B. -

Inherited Renal Tubulopathies—Challenges and Controversies
G C A T T A C G G C A T genes Review Inherited Renal Tubulopathies—Challenges and Controversies Daniela Iancu 1,* and Emma Ashton 2 1 UCL-Centre for Nephrology, Royal Free Campus, University College London, Rowland Hill Street, London NW3 2PF, UK 2 Rare & Inherited Disease Laboratory, London North Genomic Laboratory Hub, Great Ormond Street Hospital for Children National Health Service Foundation Trust, Levels 4-6 Barclay House 37, Queen Square, London WC1N 3BH, UK; [email protected] * Correspondence: [email protected]; Tel.: +44-2381204172; Fax: +44-020-74726476 Received: 11 February 2020; Accepted: 29 February 2020; Published: 5 March 2020 Abstract: Electrolyte homeostasis is maintained by the kidney through a complex transport function mostly performed by specialized proteins distributed along the renal tubules. Pathogenic variants in the genes encoding these proteins impair this function and have consequences on the whole organism. Establishing a genetic diagnosis in patients with renal tubular dysfunction is a challenging task given the genetic and phenotypic heterogeneity, functional characteristics of the genes involved and the number of yet unknown causes. Part of these difficulties can be overcome by gathering large patient cohorts and applying high-throughput sequencing techniques combined with experimental work to prove functional impact. This approach has led to the identification of a number of genes but also generated controversies about proper interpretation of variants. In this article, we will highlight these challenges and controversies. Keywords: inherited tubulopathies; next generation sequencing; genetic heterogeneity; variant classification. 1. Introduction Mutations in genes that encode transporter proteins in the renal tubule alter kidney capacity to maintain homeostasis and cause diseases recognized under the generic name of inherited tubulopathies. -

Methods in and Applications of the Sequencing of Short Non-Coding Rnas" (2013)
University of Pennsylvania ScholarlyCommons Publicly Accessible Penn Dissertations 2013 Methods in and Applications of the Sequencing of Short Non- Coding RNAs Paul Ryvkin University of Pennsylvania, [email protected] Follow this and additional works at: https://repository.upenn.edu/edissertations Part of the Bioinformatics Commons, Genetics Commons, and the Molecular Biology Commons Recommended Citation Ryvkin, Paul, "Methods in and Applications of the Sequencing of Short Non-Coding RNAs" (2013). Publicly Accessible Penn Dissertations. 922. https://repository.upenn.edu/edissertations/922 This paper is posted at ScholarlyCommons. https://repository.upenn.edu/edissertations/922 For more information, please contact [email protected]. Methods in and Applications of the Sequencing of Short Non-Coding RNAs Abstract Short non-coding RNAs are important for all domains of life. With the advent of modern molecular biology their applicability to medicine has become apparent in settings ranging from diagonistic biomarkers to therapeutics and fields angingr from oncology to neurology. In addition, a critical, recent technological development is high-throughput sequencing of nucleic acids. The convergence of modern biotechnology with developments in RNA biology presents opportunities in both basic research and medical settings. Here I present two novel methods for leveraging high-throughput sequencing in the study of short non- coding RNAs, as well as a study in which they are applied to Alzheimer's Disease (AD). The computational methods presented here include High-throughput Annotation of Modified Ribonucleotides (HAMR), which enables researchers to detect post-transcriptional covalent modifications ot RNAs in a high-throughput manner. In addition, I describe Classification of RNAs by Analysis of Length (CoRAL), a computational method that allows researchers to characterize the pathways responsible for short non-coding RNA biogenesis. -

THE NON-DYSTROPHIC MYOPATHIES JOHN PEARCE, M.B., M.R.C.P., Department of Neurology, the General Infirmary, Leeds
Postgrad Med J: first published as 10.1136/pgmj.41.476.347 on 1 June 1965. Downloaded from POSTGRAD. MED. J. (1965), 41, 347 THE NON-DYSTROPHIC MYOPATHIES JOHN PEARCE, M.B., M.R.C.P., Department of Neurology, The General Infirmary, Leeds. THE TERM 'myopathy' is applied to any disorder Polymyositis may affect people of any age, of the muscle fibre, the muscle fibre membrane, but the age of onset is from 30 to 60 in 60% the myoneural junction, or the muscle connect- of cases. The chief symptom is weakness of ive tissue. 'Non-dystrophic myopathy' includes proximal muscles of the arms and/or legs all diseases of muscle excluding those genetically in every case. Distal muscles are affected in determined primary degenerative myopathies, one third, and the neck muscles in two thirds collectively known as Muscular Dystrophy. of patients. Fever, muscular pain and tenderness TABLE 1 are seen most often in the more acute forms, CLASSIFICATION OF NON-DYSTROPHIC MYOPATHIES and their absence should not lead to neglecting 1. Inflammatory Polymyositis as a Myopathy Connective tissue disorders polymyositis possible diagnosis. Other inflammatory mvopathies Acute polymyositis is not common, but may 2. Metabolic Familial periodic paralysis progress rapidly and involve the respiratory Myopathy Muscle glycogenoses muscles, sometimes with a fatal termination Myoglobinuric myopathies within a few weeks or months. Myopathies associated with Subacute and electrolyte imbalance chronic forms are more frequent, and present Unclassified myopathies with a progressive weakness and moderate of shoulder 3. Endocrine Thyrotoxicosis wasting and pelvic girdle muscles. Myopathy Cushing's Syndrome There is sometimes no systemic disturbance, Steroid myopathy and pain and tenderness are lacking. -

A Comprehensive Network and Pathway Analysis of Human Deafness Genes
Otology & Neurotology 34:961Y970 Ó 2013, Otology & Neurotology, Inc. A Comprehensive Network and Pathway Analysis of Human Deafness Genes *Georgios A. Stamatiou and †Konstantina M. Stankovic *Department of Otolaryngology, Hippokration General Hospital, University of Athens, Athens, Greece; and ÞDepartment of Otology and Laryngology, Harvard Medical School and Department of Otolaryngology, Massachusetts Eye and Ear Infirmary, Boston, Massachusetts, U.S.A. Objective: To perform comprehensive network and pathway factor beta1 (TGFB1) for Group 1, MAPK3/MAPK1 MAP kinase analyses of the genes known to cause genetic hearing loss. (ERK 1/2) and the G protein coupled receptors (GPCR) for Study Design: In silico analysis of deafness genes using inge- Group 2, and TGFB1 and hepatocyte nuclear factor 4 alpha (HNF4A) nuity pathway analysis (IPA). for Group 3. The nodal molecules included not only those known Methods: Genes relevant for hearing and deafness were iden- to be associated with deafness (GPCR), or with predisposition to tified through PubMed literature searches and the Hereditary otosclerosis (TGFB1), but also novel genes that have not been Hearing Loss Homepage. The genes were assembled into 3 groups: described in the cochlea (HNF4A) and signaling kinases (ERK 1/2). 63 genes that cause nonsyndromic deafness, 107 genes that cause Conclusion: A number of molecules that are likely to be key nonsyndromic or syndromic sensorineural deafness, and 112 genes mediators of genetic hearing loss were identified through three associated with otic capsule development and malformations. Each different network and pathway analyses. The molecules included group of genes was analyzed using IPA to discover the most new candidate genes for deafness. -

Structural Capacitance in Protein Evolution and Human Diseases
Article Structural Capacitance in Protein Evolution and Human Diseases Chen Li 1,2, Liah V.T. Clark 1, Rory Zhang 1, Benjamin T. Porebski 1,3, Julia M. McCoey 1, Natalie A. Borg 1, Geoffrey I. Webb 4, Itamar Kass 1,5, Malcolm Buckle 6, Jiangning Song 1, Adrian Woolfson 7 and Ashley M. Buckle 1 1 - Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Clayton, Victoria 3800, Australia 2 - Department of Biology, Institute of Molecular Systems Biology, ETH Zurich, Zurich 8093, Switzerland 3 - Medical Research Council Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge, CB2 0QH, UK 4 - Faculty of Information Technology, Monash University, Clayton, Victoria 3800, Australia 5 - Amai Proteins, Prof. A. D. Bergman 2B, Suite 212, Rehovot 7670504, Israel 6 - LBPA, ENS Cachan, CNRS, Université Paris-Saclay, F-94235 Cachan, France 7 - Nouscom, Baumleingasse, CH-4051 Basel, Switzerland Correspondence to Adrian Woolfson and Ashley M. Buckle: [email protected]; [email protected] https://doi.org/10.1016/j.jmb.2018.06.051 Edited by Dan Tawfik Abstract Canonical mechanisms of protein evolution include the duplication and diversification of pre-existing folds through genetic alterations that include point mutations, insertions, deletions, and copy number amplifications, as well as post-translational modifications that modify processes such as folding efficiency and cellular localization. Following a survey of the human mutation database, we have identified an additional mechanism that we term “structural capacitance,” which results in the de novo generation of microstructure in previously disordered regions. We suggest that the potential for structural capacitance confers select proteins with the capacity to evolve over rapid timescales, facilitating saltatory evolution as opposed to gradualistic canonical Darwinian mechanisms. -

Prevalence and Incidence of Rare Diseases: Bibliographic Data
Number 1 | January 2019 Prevalence and incidence of rare diseases: Bibliographic data Prevalence, incidence or number of published cases listed by diseases (in alphabetical order) www.orpha.net www.orphadata.org If a range of national data is available, the average is Methodology calculated to estimate the worldwide or European prevalence or incidence. When a range of data sources is available, the most Orphanet carries out a systematic survey of literature in recent data source that meets a certain number of quality order to estimate the prevalence and incidence of rare criteria is favoured (registries, meta-analyses, diseases. This study aims to collect new data regarding population-based studies, large cohorts studies). point prevalence, birth prevalence and incidence, and to update already published data according to new For congenital diseases, the prevalence is estimated, so scientific studies or other available data. that: Prevalence = birth prevalence x (patient life This data is presented in the following reports published expectancy/general population life expectancy). biannually: When only incidence data is documented, the prevalence is estimated when possible, so that : • Prevalence, incidence or number of published cases listed by diseases (in alphabetical order); Prevalence = incidence x disease mean duration. • Diseases listed by decreasing prevalence, incidence When neither prevalence nor incidence data is available, or number of published cases; which is the case for very rare diseases, the number of cases or families documented in the medical literature is Data collection provided. A number of different sources are used : Limitations of the study • Registries (RARECARE, EUROCAT, etc) ; The prevalence and incidence data presented in this report are only estimations and cannot be considered to • National/international health institutes and agencies be absolutely correct. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase