The Nucleoporin Nup98 Associates with the Intranuclear Filamentous Protein Network of TPR
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Splicing Factor XAB2 Interacts with ERCC1-XPF and XPG for RNA-Loop Processing During Mammalian Development
bioRxiv preprint doi: https://doi.org/10.1101/2020.07.20.211441; this version posted July 21, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. The Splicing Factor XAB2 interacts with ERCC1-XPF and XPG for RNA-loop processing during mammalian development Evi Goulielmaki1*, Maria Tsekrekou1,2*, Nikos Batsiotos1,2, Mariana Ascensão-Ferreira3, Eleftheria Ledaki1, Kalliopi Stratigi1, Georgia Chatzinikolaou1, Pantelis Topalis1, Theodore Kosteas1, Janine Altmüller4, Jeroen A. Demmers5, Nuno L. Barbosa-Morais3, George A. Garinis1,2* 1. Institute of Molecular Biology and Biotechnology, Foundation for Research and Technology- Hellas, GR70013, Heraklion, Crete, Greece, 2. Department of Biology, University of Crete, Heraklion, Crete, Greece, 3. Instituto de Medicina Molecular João Lobo Antunes, Faculdade de Medicina da Universidade de Lisboa, Avenida Professor Egas Moniz, 1649-028 Lisboa, Portugal, 4. Cologne Center for Genomics (CCG), Institute for Genetics, University of Cologne, 50931, Cologne, Germany, 5. Proteomics Center, Netherlands Proteomics Center, and Department of Biochemistry, Erasmus University Medical Center, the Netherlands. Corresponding author: George A. Garinis ([email protected]) *: equally contributing authors bioRxiv preprint doi: https://doi.org/10.1101/2020.07.20.211441; this version posted July 21, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract RNA splicing, transcription and the DNA damage response are intriguingly linked in mammals but the underlying mechanisms remain poorly understood. Using an in vivo biotinylation tagging approach in mice, we show that the splicing factor XAB2 interacts with the core spliceosome and that it binds to spliceosomal U4 and U6 snRNAs and pre-mRNAs in developing livers. -

Nucleolus: a Central Hub for Nuclear Functions Olga Iarovaia, Elizaveta Minina, Eugene Sheval, Daria Onichtchouk, Svetlana Dokudovskaya, Sergey Razin, Yegor Vassetzky
Nucleolus: A Central Hub for Nuclear Functions Olga Iarovaia, Elizaveta Minina, Eugene Sheval, Daria Onichtchouk, Svetlana Dokudovskaya, Sergey Razin, Yegor Vassetzky To cite this version: Olga Iarovaia, Elizaveta Minina, Eugene Sheval, Daria Onichtchouk, Svetlana Dokudovskaya, et al.. Nucleolus: A Central Hub for Nuclear Functions. Trends in Cell Biology, Elsevier, 2019, 29 (8), pp.647-659. 10.1016/j.tcb.2019.04.003. hal-02322927 HAL Id: hal-02322927 https://hal.archives-ouvertes.fr/hal-02322927 Submitted on 18 Nov 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Nucleolus: A Central Hub for Nuclear Functions Olga Iarovaia, Elizaveta Minina, Eugene Sheval, Daria Onichtchouk, Svetlana Dokudovskaya, Sergey Razin, Yegor Vassetzky To cite this version: Olga Iarovaia, Elizaveta Minina, Eugene Sheval, Daria Onichtchouk, Svetlana Dokudovskaya, et al.. Nucleolus: A Central Hub for Nuclear Functions. Trends in Cell Biology, Elsevier, 2019, 29 (8), pp.647-659. 10.1016/j.tcb.2019.04.003. hal-02322927 HAL Id: hal-02322927 https://hal.archives-ouvertes.fr/hal-02322927 Submitted on 18 Nov 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. -

Unit 6-Nucleus
Unit 6-Nucleus Dr. Pallee shree Nucleus • Nucleus is the most important organelle in the cell • It distinguishes eukaryotic from prokaryotic cells • By housing the cell's genome, the nucleus serves both as the repository of genetic information and as the cell's control center • DNA replication, transcription, and RNA processing all take place within the nucleus Cont… • A nucleus is a double-membraned eukaryotic cell organelle that contains the genetic material. • It appears in an oval shape averages 5µm in width. • It often lies in the centre of a cell • The nucleus was the first organelle to be discovered • Nuclei 1st discovered and named by Robert Brown • Role of nucleus 1st demonestrated by Max Hammerling Ultra structure of Nucleus 1. Nuclear envelope 2. nuclear pores 3. Nucleoplasm 4. Nucleolus 5. Chromosomes 1. Structure of Nuclear envelope • The nuclear envelope has a complex structure consisting of a) Two nuclear membranes separated by a perinuclear space measuring about 20–40 nm across b) Underlying nuclear lamina • The nucleus is surrounded by a system of two concentric membranes, called the inner and outer nuclear membranes • The inner and outer nuclear membranes are joined at nuclear pore complexes a. Nuclear membranes • The outer nuclear membrane is continuous with the endoplasmic reticulum, so the space between the • The critical function of the inner and outer nuclear membranes nuclear membranes is to act as is directly connected with the lumen a barrier that separates the of the ER contents of the nucleus from the cytoplasm. • It is functionally similar to the membranes of the ER and has • Like other cell membranes, ribosomes bound to its cytoplasmic each nuclear membrane is a surface but protein composition phospholipid bilayer differ slightly as they are enriched in permeable only to small proteins which binds to cytoskeleton nonpolar molecules • The inner nuclear membrane carries • Other molecules are unable to proteins that are specific to the diffuse through the bilayer. -

1 the Nucleoporin ELYS Regulates Nuclear Size by Controlling NPC
bioRxiv preprint doi: https://doi.org/10.1101/510230; this version posted January 2, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. The nucleoporin ELYS regulates nuclear size by controlling NPC number and nuclear import capacity Predrag Jevtić1,4, Andria C. Schibler2,4, Gianluca Pegoraro3, Tom Misteli2,*, Daniel L. Levy1,* 1 Department of Molecular Biology, University of Wyoming, Laramie, WY, 82071 2 National Cancer Institute, NIH, Bethesda, MD, 20892 3 High Throughput Imaging Facility (HiTIF), National Cancer Institute, NIH, Bethesda, MD, 20892 4 Co-first authors *Corresponding authors: Daniel L. Levy Tom Misteli University of Wyoming National Cancer Institute, NIH Department of Molecular Biology Center for Cancer Research 1000 E. University Avenue 41 Library Drive, Bldg. 41, B610 Laramie, WY, 82071 Bethesda, MD, 20892 Phone: 307-766-4806 Phone: 240-760-6669 Fax: 307-766-5098 Fax: 301-496-4951 E-mail: [email protected] E-mail: [email protected] Running Head: ELYS is a nuclear size effector Abbreviations: NE, nuclear envelope; NPC, nuclear pore complex; ER, endoplasmic reticulum; Nup, nucleoporin; FG-Nup, phenylalanine-glycine repeat nucleoporin 1 bioRxiv preprint doi: https://doi.org/10.1101/510230; this version posted January 2, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. -

Functional Proximity Mapping of RNA Binding Proteins Uncovers a Mitochondrial Mrna Anchor That Promotes Stress Recovery
Functional proximity mapping of RNA binding proteins uncovers a mitochondrial mRNA anchor that promotes stress recovery Wei Qin Stanford University Samuel Myers Broad Institute Dominique Carey The Broad Institute of MIT and Harvard Steven Carr Broad Institute Alice Ting ( [email protected] ) Stanford University https://orcid.org/0000-0002-8277-5226 Article Keywords: Unbiased Proteome Discovery, Nucleolus, Outer Mitochondrial Membrane, Local Translation, Functional Protein Mapping Posted Date: November 12th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-103196/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/49 Abstract Proximity labeling (PL) with genetically-targeted promiscuous enzymes has emerged as a powerful tool for unbiased proteome discovery. By combining the spatiotemporal specicity of PL with methods for functional protein enrichment, it should be possible to map specic protein subclasses within distinct compartments of living cells. Here we demonstrate this capability for RNA binding proteins (RBPs), by combining peroxidase-based PL with organic-aqueous phase separation of crosslinked protein-RNA complexes (“APEX-PS”). We validated APEX-PS by mapping nuclear RBPs, then applied it to uncover the RBPomes of two unpuriable subcompartments - the nucleolus and the outer mitochondrial membrane (OMM). At the OMM, we discovered the RBP SYNJ2BP, which retains specic nuclear-encoded mitochondrial mRNAs during translation stress, to promote their local translation and import of protein products into the mitochondrion during stress recovery. APEX-PS is a versatile tool for compartment- specic RBP discovery and expands the scope of PL to functional protein mapping. Introduction RNA-protein interactions are pervasive in both transient and stable macromolecular complexes underlying transcription, translation and stress response 1, 2. -
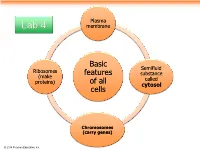
Basic Features of All Cells
Plasma Lab 4 membrane Basic Semifluid Ribosomes features substance (make called proteins) of all cytosol cells Chromosomes (carry genes) © 2014 Pearson Education, Inc. Prokaryotic cells are characterized by having . No nucleus . DNA in an unbound region called the nucleoid . No membrane-bound organelles . Cytoplasm bound by the plasma membrane © 2014 Pearson Education, Inc. Eukaryotic cells are characterized by having • DNA in a nucleus that is bounded by a membranous nuclear envelope • Membrane-bound organelles • Cytoplasm in the region between the plasma membrane and nucleus Eukaryotic cells are generally much larger than prokaryotic cells © 2014 Pearson Education, Inc. Figure 6.8a ENDOPLASMIC RETICULUM (ER) Nuclear envelope Rough ER Smooth ER Nucleolus NUCLEUS Flagellum Chromatin Centrosome Plasma membrane CYTOSKELETON: Microfilaments Intermediate filaments Microtubules Ribosomes Microvilli Golgi apparatus Peroxisome Lysosome Mitochondrion © 2014 Pearson Education, Inc. Figure 6.8b Nuclear envelope NUCLEUS Nucleolus Rough ER Chromatin Smooth ER Ribosomes Golgi Central vacuole apparatus Microfilaments CYTOSKELETON Microtubules Mitochondrion Peroxisome Plasma Chloroplast membrane Cell wall Plasmodesmata Wall of adjacent cell © 2014 Pearson Education, Inc. © 2014 Pearson Education, Inc. © 2014 Pearson Education, Inc. © 2014 Pearson Education, Inc. Table 6.1 © 2014 Pearson Education, Inc. Cell Walls of Plants The cell wall is an extracellular structure that distinguishes plant cells from animal cells Plant cell walls Prokaryotes, are made -

Driving Nucleolar Assembly
Downloaded from genesdev.cshlp.org on October 5, 2021 - Published by Cold Spring Harbor Laboratory Press PERSPECTIVE Driving nucleolar assembly Kathleen L. McCann,1 and Susan J. Baserga1,2,3,4 1Department of Genetics, 2Department of Molecular Biophysics and Biochemistry, 3Department of Therapeutic Radiology, Yale University School of Medicine, New Haven, Connecticut 06520, USA In this issue of Genes & Development,Grobandcol- breaks down (open mitosis), transcription of the ribosomal leagues (pp. 220–230) identify the minimal molecular re- RNA (rRNA) is inhibited, and the nucleolus disassem- quirements to assemble a fully functional nucleolus in bles. Upon completion of mitosis, rRNA transcription human cells and demonstrate the importance of the is reinitiated within the NOR, ribosome biogenesis fac- nucleolar transcription factor upstream binding factor tors are recruited, and the nucleolus is assembled. Only (UBF) as a mitotic bookmark at the ribosomal DNA NORs that are actively engaged in transcription can (rDNA). direct nucleolar assembly (Hernandez-Verdun 2011). Consequently, the nucleolus truly appears to be ‘‘an organelle formed by the act of building a ribosome’’ (Melese and Xue 1995). In all eukaryotes, the nucleolus is an essential, non- To begin to reveal the molecular mechanisms of nucle- membrane-bound organelle within the nucleus. Assem- olar formation, McStay’s laboratory (Mais et al. 2005; Grob bled around the ribosomal DNA (rDNA), the nucleolus et al. 2014) has applied synthetic biology. In an earlier is the site of ribosome biogenesis. In addition to its role report, they introduced 6.4 kb of DNA repeat sequences in making ribosomes, the nucleolus functions in other from the intergenic spacer of the Xenopus ribosomal gene important cellular processes, including stress sensing, into a noncanonical site in the human genome. -

Segregation of the Nucleolus Produced by Anthramycin1
[CANCER RESEARCH 28, 81-90, January 1968] Segregation of the Nucleolus Produced by Anthramycin1 Curtis Harris, Harold Grady, and Donald Svoboda Department oj Pathology, University oj Kansas Medical Center, Kansas City, Kansas 66103 SUMMARY mycin does not produce toxicity in the bone marrow and gastrointestinal tract (49). It possesses certain structural fea Anthramycin, an antibiotic possessing antitumor properties, tures in common with actinomycin D and the postulated biosyn- was administered to rats and mice in a single intraperitoneal thetic intermediates of the actinomycin^, 3-hydroxy-4-methyI- close ranging from 1 to 10 mg/kg body weight. Light micro anthraniloyl peptides (3,47). Like kanamycin, neomycin, and scopy showed that, at the highest dose, anthramycin produced other antibiotics (48), anthramycin permanently bleaches necrosis in the pancreas of rats but not in mice. By electron Euglena gracilis (11). microscopy, nucleolar changes were observed in the spleen and This report demonstrates that anthramycin is another agent kidney of both species by 1 hour. Acinar cells of the pancreas which causes nucleolar changes in pancreatic cells, reticulo showed nucleolar caps and segregation of nucleolar constituents endothelial cells of liver and spleen, and in proximal tubular in both species at 3 to 4 hours. By 10 hours segregation of the epithelium in the kidney. nucleolus and focal cytoplasmic degradation were prominent in pancreatic cells of rats given higher doses but were not MATERIALS AND METHODS observed in hepatocytes of either species. Anthramycin produced nucleolar alterations in Kupffer cells, Thirty-six inbred Fisher-344 male rats weighing between 95 endocrine and exocrine pancreatic cells, proximal tubular cells and 240 gm and fifteen C57 black mice weighing between 20 of the kidney, and in the reticuloendothelial cells of the spleen and 30 gm were used. -

Caspases Mediate Nucleoporin Cleavage, but Not Early Redistribution of Nuclear Transport Factors and Modulation of Nuclear Permeability in Apoptosis
Cell Death and Differentiation (2001) 8, 495 ± 505 ã 2001 Nature Publishing Group All rights reserved 1350-9047/01 $15.00 www.nature.com/cdd Caspases mediate nucleoporin cleavage, but not early redistribution of nuclear transport factors and modulation of nuclear permeability in apoptosis E Ferrando-May1, V Cordes2,3, I Biller-Ckovric1, J Mirkovic1, Val-Ala-aspartyl-¯uoromethylketone; DEVD-CHO, N-acetyl-Asp- DGoÈ rlich4 and P Nicotera*,5 Glu-Val-Asp-aldehyde 1 Chair of Molecular Toxicology, Department of Biology, University of Konstanz, 78457 Konstanz, Germany Introduction 2 Karolinska Institutet, Medical Nobel Institute, Department of Cellular and Molecular Biology, S-17177 Stockholm, Sweden The most evident morphological feature of apoptosis is the 3 Division of Cell Biology, Germany Cancer Research Center, D-69120, disassembly of the nucleus, which involves the condensation Heidelberg, Germany 4 of chromatin and its segregation into membrane-enclosed Zentrum fuÈr Molekulare Biologie der UniversitaÈt Heidelberg, D-69120, 1 Heidelberg, Germany particles. Biochemical hallmarks of apoptotic nuclear 5 MRC Toxicology Unit, Hodgkin Building, University of Leicester, Lancaster execution are DNA cleavage in large and small (oligonu- Road, Leicester LE1 9HN, UK cleosomal-sized) fragments, as well as the specific proteo- * Corresponding author: P Nicotera, MRC Toxicology Unit, Hodgkin Building, lysis of several nuclear substrates. Major effectors of University of Leicester, Lancaster Road, Leicester LE1 9HN, UK. apoptotic nuclear changes are members of the cysteine Tel +44-116-2525611; Fax: +44-116-2525616; E-mail: [email protected] protease family of caspases. Nuclear substrates for caspases 2,3 Received 23.11.00; revised 22.12.00; accepted 29.12.00 include nucleoskeletal elements like lamins, and proteins Edited by M Piacentini involved in the organisation and replication of DNA, like SAF- A, MCM3 and RCF140.4±6 Cleavage of nuclear proteins may have important Abstract implications for the apoptotic process. -

Novel Targets of Apparently Idiopathic Male Infertility
International Journal of Molecular Sciences Review Molecular Biology of Spermatogenesis: Novel Targets of Apparently Idiopathic Male Infertility Rossella Cannarella * , Rosita A. Condorelli , Laura M. Mongioì, Sandro La Vignera * and Aldo E. Calogero Department of Clinical and Experimental Medicine, University of Catania, 95123 Catania, Italy; [email protected] (R.A.C.); [email protected] (L.M.M.); [email protected] (A.E.C.) * Correspondence: [email protected] (R.C.); [email protected] (S.L.V.) Received: 8 February 2020; Accepted: 2 March 2020; Published: 3 March 2020 Abstract: Male infertility affects half of infertile couples and, currently, a relevant percentage of cases of male infertility is considered as idiopathic. Although the male contribution to human fertilization has traditionally been restricted to sperm DNA, current evidence suggest that a relevant number of sperm transcripts and proteins are involved in acrosome reactions, sperm-oocyte fusion and, once released into the oocyte, embryo growth and development. The aim of this review is to provide updated and comprehensive insight into the molecular biology of spermatogenesis, including evidence on spermatogenetic failure and underlining the role of the sperm-carried molecular factors involved in oocyte fertilization and embryo growth. This represents the first step in the identification of new possible diagnostic and, possibly, therapeutic markers in the field of apparently idiopathic male infertility. Keywords: spermatogenetic failure; embryo growth; male infertility; spermatogenesis; recurrent pregnancy loss; sperm proteome; DNA fragmentation; sperm transcriptome 1. Introduction Infertility is a widespread condition in industrialized countries, affecting up to 15% of couples of childbearing age [1]. It is defined as the inability to achieve conception after 1–2 years of unprotected sexual intercourse [2]. -

Characterization of Nucleoporin 98-Fusion Proteins
Department for Biomedical Sciences University of Veterinary Medicine Vienna Institute of Medical Biochemistry (Head: Prof. Dr. Florian Grebien) Characterization of Nucleoporin 98-Fusion Proteins Master’s thesis submitted for the fulfillment of the requirements for the degree of Master of Science M.Sc. University of Veterinary Medicine Vienna Submitted by Theresa Humer, B.Sc. Vienna, January 2020 SUPERVISION Internal Supervisor (University of Veterinary Medicine Vienna) Prof. Dr. Florian Grebien Institute of Medical Biochemistry SWORN DECLARATION I hereby declare that I prepared this work independently and without help from third parties, that I did not use sources other than the ones referenced and that I have indicated passages taken from those sources. This thesis was not previously submitted in identical or similar form to any other examination board, nor was it published. .................................................................... Theresa Humer Vienna, January 2020 ACKNOWLEDGEMENT First, I want to thank Dr. Florian Grebien for giving me the opportunity to work on this project and his constructive feedback to my work and thesis. I am very grateful for having the chance to write my thesis under his supervision. Furthermore, I would like to express my appreciation to Stefan Terlecki-Zaniewicz, who guided me through the whole project and supported me during my work. I want to thank all members of the Grebien group for their support, advice and warm atmosphere from the first day on. This research was supported using resources of the VetCore Facility (Imaging) of the University of Veterinary Medicine Vienna with great thanks to Ursula Reichart for fruitful discussions. Further, I would like to generally thank my friends and family for their steady support and especially to my parents, who encouraged me a lot during my years of study. -

The Combined Detection of Amphiregulin, Cyclin A1 and DDX20/Gemin3 Expression Predicts Aggressive Forms of Oral Squamous Cell Carcinoma
British Journal of Cancer www.nature.com/bjc ARTICLE OPEN Molecular Diagnostics The combined detection of Amphiregulin, Cyclin A1 and DDX20/Gemin3 expression predicts aggressive forms of oral squamous cell carcinoma 1 2 3 1 1 1 1 Ekaterina Bourova-Flin✉, Samira Derakhshan , Afsaneh✉ Goudarzi , Tao Wang , Anne-Laure Vitte , Florent Chuffart , Saadi Khochbin , Sophie Rousseaux 1 and Pouyan Aminishakib 2,4 © The Author(s) 2021 BACKGROUND: Large-scale genetic and epigenetic deregulations enable cancer cells to ectopically activate tissue-specific expression programmes. A specifically designed strategy was applied to oral squamous cell carcinomas (OSCC) in order to detect ectopic gene activations and develop a prognostic stratification test. METHODS: A dedicated original prognosis biomarker discovery approach was implemented using genome-wide transcriptomic data of OSCC, including training and validation cohorts. Abnormal expressions of silent genes were systematically detected, correlated with survival probabilities and evaluated as predictive biomarkers. The resulting stratification test was confirmed in an independent cohort using immunohistochemistry. RESULTS: A specific gene expression signature, including a combination of three genes, AREG, CCNA1 and DDX20, was found associated with high-risk OSCC in univariate and multivariate analyses. It was translated into an immunohistochemistry-based test, which successfully stratified patients of our own independent cohort. DISCUSSION: The exploration of the whole gene expression profile characterising