Philosophical Transactions of the Royal Society B Fungal
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Calonectria Ilicicola
OctoberPathogen of11 the month –October 2011 a b c d Reitsma & Fig. 1. Calonectria ilicicola; asci with ascospores (a); microsclerotia on a papaya root (b); conidium of anamorph, Cylindrocladium parasiticum (c); orange perithecia on papaya root (d); Photo credits M. Male (a,b,c), L. Vawdrey (d) Disease: Collar rot of papaya Pathogen: Calonectria ilicicola (anamorph: Cylindrocladium parasiticum) Boedijn Classification: K: Fungi, D: Ascomycota, C: Sordariomycetes, O: Hypocreales, F: Nectriaceae Calonectria ilicicola (Fig. 1) is a fungal pathogen found throughout the world infecting a wide variety of crops. It is best known as the causal agent of peg, pod and root necrosis of peanuts, collar rot of koa, leaf spot in some eucalypts and various root and collar rots of soybean, anthurium, groundnut and lucerne. In Australia, it causes black rot of peanut and collar rot of papaya. Its development is favoured by poorly drained soils. Host Range: Key Distinguishing Features: C. ilicicola is found throughout the world. It is In seedlings, the initial symptoms are of a discoloured pathogenic on a wide variety of plants including water soaked, root collar. As this rot develops, plants peanuts, soybean, anthurium, eucalypts, lucerne, are stunted and leaves become chlorotic and wilt. groundnut and papaya. Eventually, orange to red perithecia and thick-walled ilicicola brown microsclerotia will form on roots at or near the Impact: soil line (Fig 1 b,d). In culture, asci are clavate, 90- In far north Queensland, collar rot of papaya caused 140 x 12-19μm, tapering to a long thin stalk by C. ilicicola affects young plants in poorly drained containing eight hyaline ascospores (Fig1,a). -

Cylindrocladium Buxicola Nom. Cons. Prop.(Syn. Calonectria
I Promotors: Prof. dr. ir. Monica Höfte Laboratory of Phytopathology, Department of Crop Protection Faculty of Bioscience Engineering Ghent University Dr. ir. Kurt Heungens Institute for Agricultural and Fisheries Research (ILVO) Plant Sciences Unit - Crop Protection Dean: Prof. dr. ir. Guido Van Huylenbroeck Rector: Prof. dr. Anne De Paepe II Bjorn Gehesquière Cylindrocladium buxicola nom. cons. prop. (syn. Calonectria pseudonaviculata) on Buxus: molecular characterization, epidemiology, host resistance and fungicide control Thesis submitted in fulfillment of the requirements for the degree of Doctor (PhD) in Applied Biological Sciences III Dutch translation of the title: Cylindrocladium buxicola nom. cons. prop. (syn. Calonectria pseudonaviculata) in Buxus: moleculaire karakterisering, epidemiologie, waardplantresistentie en chemische bestrijding. Please refer to this work as follows: Gehesquière B. (2014). Cylindrocladium buxicola nom. cons. prop. (syn. Calonectria pseudonaviculata) on Buxus: molecular characterization, epidemiology, host resistance and fungicide control. Phd Thesis. Ghent University, Belgium The author and the promotors give authorisation to consult and to copy parts of this work for personal use only. Any other use is limited by Laws of Copyright. Permission to reproduce any material contained in this work should be obtained from the author. The promotors, The author, Prof. dr. ir. M. Höfte Dr. ir. K. Heungens ir. B. Gehesquière IV Een woordje van dank…. Dit dankwoord schrijven is ongetwijfeld het leukste onderdeel van deze thesis, en een mooie afsluiting van een interessante periode. Terugblikkend op de voorbije vier jaren kan ik enkel maar beamen dat een doctoraat zoveel meer is dan een wetenschappelijke uitdaging. Het is een levensreis in al zijn facetten, waarbij ik mezelf heb leren kennen in al mijn goede en slechte kantjes. -

Epidemiological Studies on the Infection Process and Symptom Expression of Soybean Sudden Death Syndrome Carlos Cecilio Gongora-Canul Iowa State University
Iowa State University Capstones, Theses and Graduate Theses and Dissertations Dissertations 2010 Epidemiological studies on the infection process and symptom expression of soybean sudden death syndrome Carlos Cecilio Gongora-canul Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/etd Part of the Plant Pathology Commons Recommended Citation Gongora-canul, Carlos Cecilio, "Epidemiological studies on the infection process and symptom expression of soybean sudden death syndrome" (2010). Graduate Theses and Dissertations. 11510. https://lib.dr.iastate.edu/etd/11510 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. Epidemiological studies on the infection process and symptom expression of soybean sudden death syndrome by Carlos Cecilio Góngora-Canul A dissertation submitted to the graduate faculty in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Major: Plant Pathology Program of Study Committee: Leonor Leandro, Major Professor Gary Munkvold Greg Tylka X. B Yang Dan Nordman Iowa State University Ames, Iowa 2010 Copyright © Carlos Cecilio Góngora-Canul, 2010. All rights reserved. ii DEDICATION To my Lord, for giving me the blessing and the adventure to live. To my mother Elsa and my father Elias for their endless love and to all my brothers and sisters (Javier, Roberto, Martha, Manuel, Enrique and Nicte-Há) for all their great love affection. -

Pathogenicity and Molecular Detection of Nectriaceous Fungi Associated with Black Root Rot of Avocado
IX World Avocado Congress, 23 – 27 September, 2019, Medellín, Colombia WAC-130 PATHOGENICITY AND MOLECULAR DETECTION OF NECTRIACEOUS FUNGI ASSOCIATED WITH BLACK ROOT ROT OF AVOCADO L. E. Parkinson, D. P. Le, R. G. Shivas and E. K. Dann Dr Louisamarie Parkinson BBiotech(Hons), PhD Centre for Horticultural Science Queensland Alliance for Agriculture and Food Innovation (QAAFI) The University of Queensland, Brisbane Qld 4072 Australia [email protected] | https://www.qaafi.uq.edu.au THE UNIVERSITY OF QUEENSLAND St Lucia, Brisbane, Queensland Australia PATHOGENICITY AND MOLECULAR DETECTION OF NECTRIACEOUS FUNGI ASSOCIATED WITH BLACK ROOT ROT OF AVOCADO L. E. Parkinson1, D. P. Le1, R. G. Shivas2, E. K. Dann1 1 Queensland Alliance for Agriculture and Food Innovation, The University of Queensland, Australia 2Centre for Crop Health, The University of Southern Queensland, Australia KEY WORDS Calonectria, Calonectria ilicicola, Dactylonectria, Dactylonectria macrodidyma, diagnostic test, diversity, loop-mediated isothermal amplification (LAMP) SUMMARY Black root rot of avocado associated with soilborne nectriaceous fungi is an aggressive disease of nursery trees and young orchards transplants, causing tree stunting, wilt, severe root necrosis, rapid decline and death within a year after planting. This study aimed to identify the fungal genera associated with the disease, determine the causal agents of black root rot, and develop a rapid molecular test for detection of key pathogens in avocado roots. A disease survey in all Australian growing regions collected 153 nectriaceous fungal isolates from roots of 91 symptomatic and healthy avocado trees and other hosts including peanut, papaya, blueberry, custard apple and grapevine. The fungal isolates were identified with phylogenetic analyses of ITS, β-tubulin and Histone H3 sequenced genes. -
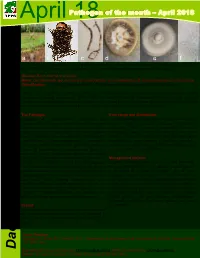
Dactylonectria Lombard & Crous
April Pathogen18 of the month – April 2018 a b c d e f Fig. 1. Black root rot symptoms in young orchard transplants (a), necrotic avocado roots (b, c) Dactylonectria macrodidyma on ½ sPDA at 3.75 cm after 10 days growth (d, e), D. macrodidyma macroconidia at 40 × magnification (f) Disease: Black root rot of avocado Name: Dactylonectria spp. including D. macrodidyma, D. novozelandica, D. pauciseptata and D. anthuriicola Classification: K: Fungi, D: Ascomycota, C: Sordariomycetes, O: Hypocreales, F: Nectriaceae Black root rot caused by nectriaceous fungi is a severe disease of avocado nursery trees and young orchard transplants, causing decline and death within one year of planting. Symptoms include stunting, wilt, leaf chlorosis and browning, leaf drop prior to tree death caused by severe necrosis of the root system. In Australia black root rot of avocado is caused by Calonectria ilicicola and several Dactylonectria spp. The Pathogen: Host range and distribution: Species of Dactylonectria (reported as Dactylonectria spp. cause root rot diseases in various Cylindrocarpon in older literature) have often been hosts including avocado (Persea americana), isolated from necrotic avocado roots. Dactylonectria grapevine (Vitis vinifera), cherimoya (Annona macrodidyma is the most prevalent of the pathogens cherimola), kiwifruit (Actinidia deliciosa) and olive found in symptomatic avocado roots. Dactylonectria (Olea europaea). Dactylonectria spp. associated with novozelandica, D. pauciseptata and D. anthuriicola avocado have been reported in Australia and Italy. have also been isolated from avocado roots and However the fungal genus is reported globally across Lombard & Crous shown to be pathogenic in glasshouse tests with numerous horticultural industries. seedlings. While Dactylonectria spp. -

Investigating Soilborne Nectriaceous Fungi Impacting Avocado Tree
Investigating soilborne nectriaceous fungi impacting avocado tree establishment in Australia Louisamarie Elicano Parkinson Bachelor of Biotechnology (Honours Class 1) A thesis submitted for the degree of Doctor of Philosophy at The University of Queensland in 2017 Queensland Alliance for Agriculture and Food Innovation 1 Abstract Black root rot is a severe disease of nursery avocado trees and orchard transplants caused by soilborne fungal pathogens in the Nectriaceae family. The genera reported to be associated with black root rot are Calonectria, Cylindrocladiella, Dactylonectria, Gliocladiopsis and Ilyonectria. These genera have not been widely studied in avocado, although the disease causes significant commercial loss, with symptoms including black, rotten roots; tree stunting; leaf wilt; and rapid tree decline and death. This PhD research aims to i) identify the nectriaceous fungal species found in avocado roots in Australia, using morphological studies and molecular phylogenetic analyses of fungal gene sequences; ii) to perform pathogenicity tests on avocado seedlings and fruit to determine the pathogenic species; iii) to investigate whether the pathogens produce phytotoxic exudates which induce and facilitate disease symptom development; iv) and to use the generated gene sequence data to develop a molecular diagnostic for rapidly detecting the pathogens. Fungal isolates were obtained from symptomatic roots from sick and healthy avocado trees, nursery stock, young orchard transplants and mature established orchard trees from all growing regions in Australia, and from other host species. Bayesian inference and Maximum likelihood phylogenetic analyses of concatenated ITS, β-tubulin and histone H3 gene loci were used to identify and classify 153 Nectriaceae isolates in the genera Calonectria, Cylindrocladiella, Dactylonectria, Gliocladiopsis, Ilyonectria and Mariannaea. -

Novel Species of Gliocladiopsis (Nectriaceae, Hypocreales, Ascomycota) from Avocado Roots (Persea Americana) in Australia
mycoscience 58 (2017) 95e102 Available online at www.sciencedirect.com journal homepage: www.elsevier.com/locate/myc Full paper Novel species of Gliocladiopsis (Nectriaceae, Hypocreales, Ascomycota) from avocado roots (Persea americana) in Australia * Louisamarie E. Parkinson a, Roger G. Shivas b, Elizabeth K. Dann a, a Queensland Alliance for Agriculture and Food Innovation, The University of Queensland, Ecosciences Precinct, 41 Boggo Road, Dutton Park, QLD 4102, Australia b Plant Pathology Herbarium, Department of Agriculture and Fisheries, Ecosciences Precinct, 41 Boggo Road, Dutton Park, QLD 4102, Australia article info abstract Article history: Root rot of avocado (Persea americana) is an important disease in seedling nurseries as well Received 24 June 2016 as in the field in eastern and southern Australia. During an investigation into the causal Received in revised form organisms of avocado root rot, 19 isolates of Gliocladiopsis were obtained from necrotic 27 October 2016 lesions on avocado roots and examined by morphology and comparison of DNA sequences Accepted 30 October 2016 from three gene loci (the internal transcribed spacer region of the nuclear rDNA, Histone Available online 7 December 2016 H3 and b-tubulin). Three new species of Gliocladiopsis are described as a result of phylo- genetic analysis of these data. One of the new species, G. peggii, formed a monophyletic Keywords: group that may represent an unresolved species complex as it contained a polytomy that Lauraceae included a well-supported clade comprising two subclades. Gliocladiopsis peggii is sister to Phylogeny G. mexicana, which is known from soil in Mexico. The remaining two new species, G. whileyi Rhizosphere and G. -

University of Catania
UNIVERSITY OF CATANIA DEPARTMENT OF AGROFOOD AND ENVIRONMENTAL MANAGEMENT SYSTEMS INTERNATIONAL PhD PLANT HEALTH TECHNOLOGIES AND PROTECTION OF AGROECOSYSTEMS CYCLE XXV 2010-2012 Detection of new Calonectria spp. and Calonectria Diseases and Changes in Fungicide Sensitivity in Calonectria scoparia Complex This thesis is presented for the degree of Doctor of Philosophy by VLADIMIRO GUARNACCIA COORDINATOR SUPERVISOR PROF. C. RAPISARDA PROF. G.POLIZZI CHAPTER 1 - The genus Calonectria and the fungicide resistance.......................... 1 1.1 Introduction............................................................................................................ 2 1.1.1 Calonectria...................................................................................................... 2 1.1.2 Importance of Calonectria.............................................................................. 3 1.1.3 Morphology..................................................................................................... 6 1.1.4 Pathogenicity................................................................................................... 9 1.1.5 Microsclerotia ................................................................................................. 9 1.1.6 Mating compatibility..................................................................................... 10 1.1.7 Phylogeny...................................................................................................... 12 1.1.7.1 Calonectria scoparia species complex ................................................. -

Species Concepts in Calonectria (Cylindrocladium)
available online at www.studiesinmycology.org StudieS in Mycology 66: 1–14. 2010. doi:10.3114/sim.2010.66.01 species concepts in Calonectria (Cylindrocladium) L. Lombard1*, P.W. Crous2, B.D. Wingfield3 and M.J. Wingfield1 1Department of Microbiology and Plant Pathology, Tree Protection Co-operative Programme, Forestry and Agricultural Biotechnology Institute, University of Pretoria, Pretoria 0002, South Africa; 2CBS-KNAW Fungal Biodiversity Centre, Uppsalalaan 8, 3584 CT Utrecht, The Netherlands; 3Department of Genetics, Forestry and Agricultural Biotechnology Institute, University of Pretoria, Pretoria 0002, South Africa *Correspondence: Lorenzo Lombard, [email protected] Abstract: Species of Calonectria and their Cylindrocladium anamorphs are important plant pathogens worldwide. At present 52 Cylindrocladium spp. and 37 Calonectria spp. are recognised based on sexual compatibility, morphology and phylogenetic inference. The polyphasic approach of integrating Biological, Morphological and Phylogenetic Species Concepts has revolutionised the taxonomy of fungi. This review aims to present an overview of published research on the genera Calonectria and Cylindrocladium as they pertain to their taxonomic history. The nomenclature as well as future research necessary for this group of fungi are also briefly discussed. Key words: Calonectria, Cylindrocladium, species concepts, nomenclature, pathogenicity. INtroductIoN topic is not treated other than in the manner in which it applies to Calonectria. The genus Calonectria (Ca.) was -

AR TICLE Lessons Learned from Moving to One
IMA FUNGUS · VOLUME 5 · A%/BB$DE./%F/B/%%/ [ ARTICLE #G <HH=K<L#9#G<%/5//O#&O&H./P/BK<#S9A#GT & ! With the changes implemented in the International Code of Nomenclature for algae, fungi and plants, fungi "#$ &[#[ Clonostachys * Nomenclature *6*[ V [&* V= !U * &&[ K !*+ (AEE*E)#Clonostachys <APH./%FS#A.5H./%FSVA%8X./%F INTRODUCTION [ # With the changes implemented in the International Code #*& of Nomenclature for algae, fungi and plants (ICN; McNeill [6 et al. 2012), fungi may no longer have more than one Chalara [!" that “…for a taxon of non- fraxinea 7 et al .//8 9 lichen-forming Ascomycota and Basidiomycota… [all names] *&<* compete for priority” regardless of their particular morph & [ Hymenoscyphus #$%#&only albidus [ H. pseudoalbidus (Queloz & !" [ et al. ./%%# [ * '& * & * ' !" principle of priority does not contribute to the nomenclatural [Chalara stability of fungi, thus exceptions can and should be made to fraxinea .//8 H. pseudoalbidus 2011, must become '[ #[ species? in Hypocreales (Rossman et al. 2013) and Leotiomycetes + (Johnston et al. 2014), I have noticed a number of issues, * * !" species of Chalara is C. fusidioides + Hymenoscyphus is H. fructigenus= [!"[ of these type species, one sees that C. fusidioides and H. The Code Decoded: a user’s guide to fructigenus the International Code of Nomenclature for algae, fungi, and presented by Réblová et al. ./%% plants./%5&!" in one genus, then most Leotiomycetes [ Chalara and Hymenoscyphus ' [ sexual state and others for one or more asexual states, one 6>@et al. (2012) © 2014 International Mycological Association You are free to share - to copy, distribute and transmit the work, under the following conditions: Attribution: [ Non-commercial: No derivative works: For any reuse or distribution, you must make clear to others the license terms of this work, which can be found at http://creativecommons.org/licenses/by-nc-nd/3.0/legalcode. -

Consortia of Anti-Nematode Fungi and Bacteria in the Rhizosphere Of
bioRxiv preprint doi: https://doi.org/10.1101/332403; this version posted May 28, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Consortia of anti-nematode fungi and bacteria in the 2 rhizosphere of soybean plants attacked by root-knot 3 nematodes 4 5 Hirokazu Toju1,2* and Yu Tanaka2,3 6 7 1Center for Ecological Research, Kyoto University, Otsu, Shiga 520-2133, Japan 8 2Precursory Research for Embryonic Science and Technology (PRESTO), Japan Science and 9 Technology Agency, Kawaguchi, Saitama 332-0012, Japan 10 3Graduate School of Agriculture, Kyoto University, Kitashirakawa-oiwake-cho, Sakyo, Kyoto, 11 606-8502, Japan 12 13 *Correspondence: Hirokazu Toju ([email protected]). 14 15 This article includes 5 Figures, 4 Tables, 5 Supplementary Figures, and 5 Supplementary 16 Data. 17 Running head: Anti-nematode microbiomes 18 bioRxiv preprint doi: https://doi.org/10.1101/332403; this version posted May 28, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 19 Abstract. 20 Cyst and root-knot nematodes are major risk factors of agroecosystem management, often 21 causing devastating impacts on crop production. The use of microbes that parasitize or prey 22 on nematodes has been considered as a promising approach for suppressing phytopathogenic 23 nematode populations. -

Species Concepts in Calonectria (Cylindrocladium)
available online at www.studiesinmycology.org StudieS in Mycology 66: 1–14. 2010. doi:10.3114/sim.2010.66.01 species concepts in Calonectria (Cylindrocladium) L. Lombard1*, P.W. Crous2, B.D. Wingfield3 and M.J. Wingfield1 1Department of Microbiology and Plant Pathology, Tree Protection Co-operative Programme, Forestry and Agricultural Biotechnology Institute, University of Pretoria, Pretoria 0002, South Africa; 2CBS-KNAW Fungal Biodiversity Centre, Uppsalalaan 8, 3584 CT Utrecht, The Netherlands; 3Department of Genetics, Forestry and Agricultural Biotechnology Institute, University of Pretoria, Pretoria 0002, South Africa *Correspondence: Lorenzo Lombard, [email protected] Abstract: Species of Calonectria and their Cylindrocladium anamorphs are important plant pathogens worldwide. At present 52 Cylindrocladium spp. and 37 Calonectria spp. are recognised based on sexual compatibility, morphology and phylogenetic inference. The polyphasic approach of integrating Biological, Morphological and Phylogenetic Species Concepts has revolutionised the taxonomy of fungi. This review aims to present an overview of published research on the genera Calonectria and Cylindrocladium as they pertain to their taxonomic history. The nomenclature as well as future research necessary for this group of fungi are also briefly discussed. Key words: Calonectria, Cylindrocladium, species concepts, nomenclature, pathogenicity. INtroductIoN topic is not treated other than in the manner in which it applies to Calonectria. The genus Calonectria (Ca.) was