<I>Gliocladiopsis Wuhanensis</I> Sp. Nov. from China
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Neural Mechanisms of Navigation Involving Interactions of Cortical and Subcortical Structures
J Neurophysiol 119: 2007–2029, 2018. First published February 14, 2018; doi:10.1152/jn.00498.2017. REVIEW Where Are You Going? The Neurobiology of Navigation Neural mechanisms of navigation involving interactions of cortical and subcortical structures James R. Hinman, Holger Dannenberg, Andrew S. Alexander, and Michael E. Hasselmo Center for Systems Neuroscience, Boston University, Boston, Massachusetts Submitted 5 July 2017; accepted in final form 1 February 2018 Hinman JR, Dannenberg H, Alexander AS, Hasselmo ME. Neural mecha- nisms of navigation involving interactions of cortical and subcortical structures. J Neurophysiol 119: 2007–2029, 2018. First published February 14, 2018; doi: 10.1152/jn.00498.2017.—Animals must perform spatial navigation for a range of different behaviors, including selection of trajectories toward goal locations and foraging for food sources. To serve this function, a number of different brain regions play a role in coding different dimensions of sensory input important for spatial behavior, including the entorhinal cortex, the retrosplenial cortex, the hippocampus, and the medial septum. This article will review data concerning the coding of the spatial aspects of animal behavior, including location of the animal within an environment, the speed of movement, the trajectory of movement, the direction of the head in the environment, and the position of barriers and objects both relative to the animal’s head direction (egocentric) and relative to the layout of the environment (allocentric). The mechanisms for coding these important spatial representations are not yet fully understood but could involve mechanisms including integration of self-motion information or coding of location based on the angle of sensory features in the environment. -

Calonectria Ilicicola
OctoberPathogen of11 the month –October 2011 a b c d Reitsma & Fig. 1. Calonectria ilicicola; asci with ascospores (a); microsclerotia on a papaya root (b); conidium of anamorph, Cylindrocladium parasiticum (c); orange perithecia on papaya root (d); Photo credits M. Male (a,b,c), L. Vawdrey (d) Disease: Collar rot of papaya Pathogen: Calonectria ilicicola (anamorph: Cylindrocladium parasiticum) Boedijn Classification: K: Fungi, D: Ascomycota, C: Sordariomycetes, O: Hypocreales, F: Nectriaceae Calonectria ilicicola (Fig. 1) is a fungal pathogen found throughout the world infecting a wide variety of crops. It is best known as the causal agent of peg, pod and root necrosis of peanuts, collar rot of koa, leaf spot in some eucalypts and various root and collar rots of soybean, anthurium, groundnut and lucerne. In Australia, it causes black rot of peanut and collar rot of papaya. Its development is favoured by poorly drained soils. Host Range: Key Distinguishing Features: C. ilicicola is found throughout the world. It is In seedlings, the initial symptoms are of a discoloured pathogenic on a wide variety of plants including water soaked, root collar. As this rot develops, plants peanuts, soybean, anthurium, eucalypts, lucerne, are stunted and leaves become chlorotic and wilt. groundnut and papaya. Eventually, orange to red perithecia and thick-walled ilicicola brown microsclerotia will form on roots at or near the Impact: soil line (Fig 1 b,d). In culture, asci are clavate, 90- In far north Queensland, collar rot of papaya caused 140 x 12-19μm, tapering to a long thin stalk by C. ilicicola affects young plants in poorly drained containing eight hyaline ascospores (Fig1,a). -

Cylindrocladium Buxicola Nom. Cons. Prop.(Syn. Calonectria
I Promotors: Prof. dr. ir. Monica Höfte Laboratory of Phytopathology, Department of Crop Protection Faculty of Bioscience Engineering Ghent University Dr. ir. Kurt Heungens Institute for Agricultural and Fisheries Research (ILVO) Plant Sciences Unit - Crop Protection Dean: Prof. dr. ir. Guido Van Huylenbroeck Rector: Prof. dr. Anne De Paepe II Bjorn Gehesquière Cylindrocladium buxicola nom. cons. prop. (syn. Calonectria pseudonaviculata) on Buxus: molecular characterization, epidemiology, host resistance and fungicide control Thesis submitted in fulfillment of the requirements for the degree of Doctor (PhD) in Applied Biological Sciences III Dutch translation of the title: Cylindrocladium buxicola nom. cons. prop. (syn. Calonectria pseudonaviculata) in Buxus: moleculaire karakterisering, epidemiologie, waardplantresistentie en chemische bestrijding. Please refer to this work as follows: Gehesquière B. (2014). Cylindrocladium buxicola nom. cons. prop. (syn. Calonectria pseudonaviculata) on Buxus: molecular characterization, epidemiology, host resistance and fungicide control. Phd Thesis. Ghent University, Belgium The author and the promotors give authorisation to consult and to copy parts of this work for personal use only. Any other use is limited by Laws of Copyright. Permission to reproduce any material contained in this work should be obtained from the author. The promotors, The author, Prof. dr. ir. M. Höfte Dr. ir. K. Heungens ir. B. Gehesquière IV Een woordje van dank…. Dit dankwoord schrijven is ongetwijfeld het leukste onderdeel van deze thesis, en een mooie afsluiting van een interessante periode. Terugblikkend op de voorbije vier jaren kan ik enkel maar beamen dat een doctoraat zoveel meer is dan een wetenschappelijke uitdaging. Het is een levensreis in al zijn facetten, waarbij ik mezelf heb leren kennen in al mijn goede en slechte kantjes. -

Epidemiological Studies on the Infection Process and Symptom Expression of Soybean Sudden Death Syndrome Carlos Cecilio Gongora-Canul Iowa State University
Iowa State University Capstones, Theses and Graduate Theses and Dissertations Dissertations 2010 Epidemiological studies on the infection process and symptom expression of soybean sudden death syndrome Carlos Cecilio Gongora-canul Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/etd Part of the Plant Pathology Commons Recommended Citation Gongora-canul, Carlos Cecilio, "Epidemiological studies on the infection process and symptom expression of soybean sudden death syndrome" (2010). Graduate Theses and Dissertations. 11510. https://lib.dr.iastate.edu/etd/11510 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. Epidemiological studies on the infection process and symptom expression of soybean sudden death syndrome by Carlos Cecilio Góngora-Canul A dissertation submitted to the graduate faculty in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Major: Plant Pathology Program of Study Committee: Leonor Leandro, Major Professor Gary Munkvold Greg Tylka X. B Yang Dan Nordman Iowa State University Ames, Iowa 2010 Copyright © Carlos Cecilio Góngora-Canul, 2010. All rights reserved. ii DEDICATION To my Lord, for giving me the blessing and the adventure to live. To my mother Elsa and my father Elias for their endless love and to all my brothers and sisters (Javier, Roberto, Martha, Manuel, Enrique and Nicte-Há) for all their great love affection. -

Pathogenicity and Molecular Detection of Nectriaceous Fungi Associated with Black Root Rot of Avocado
IX World Avocado Congress, 23 – 27 September, 2019, Medellín, Colombia WAC-130 PATHOGENICITY AND MOLECULAR DETECTION OF NECTRIACEOUS FUNGI ASSOCIATED WITH BLACK ROOT ROT OF AVOCADO L. E. Parkinson, D. P. Le, R. G. Shivas and E. K. Dann Dr Louisamarie Parkinson BBiotech(Hons), PhD Centre for Horticultural Science Queensland Alliance for Agriculture and Food Innovation (QAAFI) The University of Queensland, Brisbane Qld 4072 Australia [email protected] | https://www.qaafi.uq.edu.au THE UNIVERSITY OF QUEENSLAND St Lucia, Brisbane, Queensland Australia PATHOGENICITY AND MOLECULAR DETECTION OF NECTRIACEOUS FUNGI ASSOCIATED WITH BLACK ROOT ROT OF AVOCADO L. E. Parkinson1, D. P. Le1, R. G. Shivas2, E. K. Dann1 1 Queensland Alliance for Agriculture and Food Innovation, The University of Queensland, Australia 2Centre for Crop Health, The University of Southern Queensland, Australia KEY WORDS Calonectria, Calonectria ilicicola, Dactylonectria, Dactylonectria macrodidyma, diagnostic test, diversity, loop-mediated isothermal amplification (LAMP) SUMMARY Black root rot of avocado associated with soilborne nectriaceous fungi is an aggressive disease of nursery trees and young orchards transplants, causing tree stunting, wilt, severe root necrosis, rapid decline and death within a year after planting. This study aimed to identify the fungal genera associated with the disease, determine the causal agents of black root rot, and develop a rapid molecular test for detection of key pathogens in avocado roots. A disease survey in all Australian growing regions collected 153 nectriaceous fungal isolates from roots of 91 symptomatic and healthy avocado trees and other hosts including peanut, papaya, blueberry, custard apple and grapevine. The fungal isolates were identified with phylogenetic analyses of ITS, β-tubulin and Histone H3 sequenced genes. -

Interpreting Zheng Chenggong: the Politics of Dramatizing
, - 'I ., . UN1VERSIlY OF HAWAII UBRARY 3~31 INTERPRETING ZHENG CHENGGONG: THE POLITICS OF DRAMATIZING A HISTORICAL FIGURE IN JAPAN, CHINA, AND TAIWAN (1700-1963) A THESIS SUBMITTED TO THE GRADUATE DIVISION OF THE UNIVERSITY OF HAW AI'I IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF MASTER OF ARTS IN THEATRE AUGUST 2007 By Chong Wang Thesis Committee: Julie A. Iezzi, Chairperson Lurana D. O'Malley Elizabeth Wichmann-Walczak · - ii .' --, L-' ~ J HAWN CB5 \ .H3 \ no. YI,\ © Copyright 2007 By Chong Wang We certity that we have read this thesis and that, in our opinion, it is satisfactory in scope and quality as a thesis for the degree of Master of Arts in Theatre. TIIESIS COMMITTEE Chairperson iii ACKNOWLEDGEMENTS I want to give my wannest thanks to my family for their strong support. I also want to give my since're thanks to Dr. Julie Iezzi for her careful guidance and tremendous patience during each stage of the writing process. Finally, I want to thank my proofreaders, Takenouchi Kaori and Vance McCoy, without whom this thesis could not have been completed. - . iv ABSTRACT Zheng Chenggong (1624 - 1662) was sired by Chinese merchant-pirate in Hirado, Nagasaki Prefecture, Japan. A general at the end of the Chinese Ming Dynasty, he was a prominent leader of the movement opposing the Manchu Qing Dynasty, and in recovering Taiwan from Dutch colonial occupation in 1661. Honored as a hero in Japan, China, and Taiwan, he has been dramatized in many plays in various theatre forms in Japan (since about 1700), China (since 1906), and Taiwan (since the 1920s). -
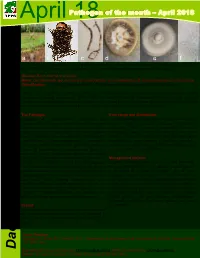
Dactylonectria Lombard & Crous
April Pathogen18 of the month – April 2018 a b c d e f Fig. 1. Black root rot symptoms in young orchard transplants (a), necrotic avocado roots (b, c) Dactylonectria macrodidyma on ½ sPDA at 3.75 cm after 10 days growth (d, e), D. macrodidyma macroconidia at 40 × magnification (f) Disease: Black root rot of avocado Name: Dactylonectria spp. including D. macrodidyma, D. novozelandica, D. pauciseptata and D. anthuriicola Classification: K: Fungi, D: Ascomycota, C: Sordariomycetes, O: Hypocreales, F: Nectriaceae Black root rot caused by nectriaceous fungi is a severe disease of avocado nursery trees and young orchard transplants, causing decline and death within one year of planting. Symptoms include stunting, wilt, leaf chlorosis and browning, leaf drop prior to tree death caused by severe necrosis of the root system. In Australia black root rot of avocado is caused by Calonectria ilicicola and several Dactylonectria spp. The Pathogen: Host range and distribution: Species of Dactylonectria (reported as Dactylonectria spp. cause root rot diseases in various Cylindrocarpon in older literature) have often been hosts including avocado (Persea americana), isolated from necrotic avocado roots. Dactylonectria grapevine (Vitis vinifera), cherimoya (Annona macrodidyma is the most prevalent of the pathogens cherimola), kiwifruit (Actinidia deliciosa) and olive found in symptomatic avocado roots. Dactylonectria (Olea europaea). Dactylonectria spp. associated with novozelandica, D. pauciseptata and D. anthuriicola avocado have been reported in Australia and Italy. have also been isolated from avocado roots and However the fungal genus is reported globally across Lombard & Crous shown to be pathogenic in glasshouse tests with numerous horticultural industries. seedlings. While Dactylonectria spp. -

Ideophones in Middle Chinese
KU LEUVEN FACULTY OF ARTS BLIJDE INKOMSTSTRAAT 21 BOX 3301 3000 LEUVEN, BELGIË ! Ideophones in Middle Chinese: A Typological Study of a Tang Dynasty Poetic Corpus Thomas'Van'Hoey' ' Presented(in(fulfilment(of(the(requirements(for(the(degree(of(( Master(of(Arts(in(Linguistics( ( Supervisor:(prof.(dr.(Jean=Christophe(Verstraete((promotor)( ( ( Academic(year(2014=2015 149(431(characters Abstract (English) Ideophones in Middle Chinese: A Typological Study of a Tang Dynasty Poetic Corpus Thomas Van Hoey This M.A. thesis investigates ideophones in Tang dynasty (618-907 AD) Middle Chinese (Sinitic, Sino- Tibetan) from a typological perspective. Ideophones are defined as a set of words that are phonologically and morphologically marked and depict some form of sensory image (Dingemanse 2011b). Middle Chinese has a large body of ideophones, whose domains range from the depiction of sound, movement, visual and other external senses to the depiction of internal senses (cf. Dingemanse 2012a). There is some work on modern variants of Sinitic languages (cf. Mok 2001; Bodomo 2006; de Sousa 2008; de Sousa 2011; Meng 2012; Wu 2014), but so far, there is no encompassing study of ideophones of a stage in the historical development of Sinitic languages. The purpose of this study is to develop a descriptive model for ideophones in Middle Chinese, which is compatible with what we know about them cross-linguistically. The main research question of this study is “what are the phonological, morphological, semantic and syntactic features of ideophones in Middle Chinese?” This question is studied in terms of three parameters, viz. the parameters of form, of meaning and of use. -

Download File
On the Periphery of a Great “Empire”: Secondary Formation of States and Their Material Basis in the Shandong Peninsula during the Late Bronze Age, ca. 1000-500 B.C.E Minna Wu Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate School of Arts and Sciences COLUMIBIA UNIVERSITY 2013 @2013 Minna Wu All rights reserved ABSTRACT On the Periphery of a Great “Empire”: Secondary Formation of States and Their Material Basis in the Shandong Peninsula during the Late Bronze-Age, ca. 1000-500 B.C.E. Minna Wu The Shandong region has been of considerable interest to the study of ancient China due to its location in the eastern periphery of the central culture. For the Western Zhou state, Shandong was the “Far East” and it was a vast region of diverse landscape and complex cultural traditions during the Late Bronze-Age (1000-500 BCE). In this research, the developmental trajectories of three different types of secondary states are examined. The first type is the regional states established by the Zhou court; the second type is the indigenous Non-Zhou states with Dong Yi origins; the third type is the states that may have been formerly Shang polities and accepted Zhou rule after the Zhou conquest of Shang. On the one hand, this dissertation examines the dynamic social and cultural process in the eastern periphery in relation to the expansion and colonization of the Western Zhou state; on the other hand, it emphasizes the agency of the periphery during the formation of secondary states by examining how the polities in the periphery responded to the advances of the Western Zhou state and how local traditions impacted the composition of the local material assemblage which lay the foundation for the future prosperity of the regional culture. -

Thank You to All of Our Reviewers in 2020
EDITORIAL https://doi.org/10.1038/s42003-021-01719-9 OPEN Thank you to all of our reviewers in 2020 5314 individual reviewers contributed to the peer review process at Communications Biology in 2020. We acknowledge and thank each of them here. Communications Biology received more than 3000 research and review article submissions last year, of which over 1700 were sent for external review. In all, 5314 individuals gave their time to ensure a fair and thorough peer review process in the uniquely challenging year that was 2020. These individuals are recognized below in alphabetical order by surname. We are truly grateful to all of our reviewers. Journal staff take full responsibility for any misspellings, omissions, or other errors. Anna M. Åsman Muneeb Ahmed Donát Alpár Luiza Aparecido Adam Abate Warish Ahmed Ahmad Alqudah Samuel Aparicio Cory Abate-Shen Bumsoo Ahn Myriam Altamirano- Sebastien Apcher Juliya Abbasi Velma Aho Bustamante Brian Appleby Geoffrey Abbott Gaurav Ahuja Matthias Altmeyer Rajendra Apte Karen Abbott Nicole Aiello-Couzo Barry Alto Catalina Arévalo Ferro 1234567890():,; Basem Abdallah Ajoy Ajoy Basak Douglas Altshuler Maria Aragona Omar Abdel-Wahab Takashi Akagi Lucia Altucci Mariaceleste Aragona Sarki Abdulkadir Ali Akbari Fernando Alvarez Midori Arai Maha Abdullah Mohsen Akbari Joao Miguel Alves Kazuharu Arakawa Ikuro Abe Fulya Akcimen Nuno Alves Beatrice Aramini Masato Abe Seiji Akimoto Marina Alves Amaral Beatriz Aranda Ethan Abel Aleksei Aksimentiev Kaushalya Amarasinghe Zolt Arany Oscar Abilez David Alabadi Ivano -

Investigating Soilborne Nectriaceous Fungi Impacting Avocado Tree
Investigating soilborne nectriaceous fungi impacting avocado tree establishment in Australia Louisamarie Elicano Parkinson Bachelor of Biotechnology (Honours Class 1) A thesis submitted for the degree of Doctor of Philosophy at The University of Queensland in 2017 Queensland Alliance for Agriculture and Food Innovation 1 Abstract Black root rot is a severe disease of nursery avocado trees and orchard transplants caused by soilborne fungal pathogens in the Nectriaceae family. The genera reported to be associated with black root rot are Calonectria, Cylindrocladiella, Dactylonectria, Gliocladiopsis and Ilyonectria. These genera have not been widely studied in avocado, although the disease causes significant commercial loss, with symptoms including black, rotten roots; tree stunting; leaf wilt; and rapid tree decline and death. This PhD research aims to i) identify the nectriaceous fungal species found in avocado roots in Australia, using morphological studies and molecular phylogenetic analyses of fungal gene sequences; ii) to perform pathogenicity tests on avocado seedlings and fruit to determine the pathogenic species; iii) to investigate whether the pathogens produce phytotoxic exudates which induce and facilitate disease symptom development; iv) and to use the generated gene sequence data to develop a molecular diagnostic for rapidly detecting the pathogens. Fungal isolates were obtained from symptomatic roots from sick and healthy avocado trees, nursery stock, young orchard transplants and mature established orchard trees from all growing regions in Australia, and from other host species. Bayesian inference and Maximum likelihood phylogenetic analyses of concatenated ITS, β-tubulin and histone H3 gene loci were used to identify and classify 153 Nectriaceae isolates in the genera Calonectria, Cylindrocladiella, Dactylonectria, Gliocladiopsis, Ilyonectria and Mariannaea. -

Download Author Version (PDF)
RSC Advances This is an Accepted Manuscript, which has been through the Royal Society of Chemistry peer review process and has been accepted for publication. Accepted Manuscripts are published online shortly after acceptance, before technical editing, formatting and proof reading. Using this free service, authors can make their results available to the community, in citable form, before we publish the edited article. This Accepted Manuscript will be replaced by the edited, formatted and paginated article as soon as this is available. You can find more information about Accepted Manuscripts in the Information for Authors. Please note that technical editing may introduce minor changes to the text and/or graphics, which may alter content. The journal’s standard Terms & Conditions and the Ethical guidelines still apply. In no event shall the Royal Society of Chemistry be held responsible for any errors or omissions in this Accepted Manuscript or any consequences arising from the use of any information it contains. www.rsc.org/advances Page 1 of 7 RSC Advances Journal Name RSC Publishing ARTICLE A new polymer with low dielectric constant based on trifluoromethyl substituted arene: preparation and Cite this: DOI: 10.1039/x0xx00000x properties Jiajia Wang, Kaikai Jin, Fengkai He, Jing Sun and Qiang Fang * Received 00th January 2012, Accepted 00th January 2012 A new polymer with low dielectric constant is reported here. This polymer contains a DOI: 10.1039/x0xx00000x trifluoromethyl substituted phenyl unit and a binaphthyl unit, and shows high thermostability with a glass translation temperature of 244 °C and a 5 wt% loss temperature at 518 °C in www.rsc.org/ nitrogen.