Overlapping Motifs on the Herpes Viral Proteins ICP27 and ORF57 Mediate
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

News & Views Research
NEWS & VIEWS RESEARCH may exhibit some tissue specificity in humans. Upstream intron Downstream intron a Sibley et al. found that genes with long introns sequence sequence tend to be expressed in the human nervous Precursor RNA system, and they identified recursively spliced 3' Splice 5' Splice RNAs expressed in the human brain6. Duff site site First step et al. detected some selectivity for recursive splicing in the brain in a screen of 20 human tissues (including fetal brain and adult cerebel- Second step lum), but this may partly reflect the difficulty of detecting recursively spliced RNAs in tis- Mature mRNA sues that express such RNAs at low levels. It will be important to determine whether this b RS exon specificity, if real, results from the tendency of recursively spliced genes to be expressed in the brain, or whether cells in the nervous system have factors that promote recursive Competing 5' splice sites First step splicing. Many genes that have long introns, including those that undergo recursive splicing, are linked to neurological diseases and to Second step 9–11 Second step autism . Whether these conditions are sometimes triggered by errors in the multi- step recursive RNA-splicing process will be an exciting avenue for future studies. ■ NMD Heidi Cook-Andersen and Miles F. Wilkinson are in the Department of Reproductive Medicine, University of Figure 1 | Mechanisms of recursive splicing. a, In recursive splicing, long intron sequences of precursor California, San Diego, La Jolla, California, RNA are removed in a stepwise process mediated by juxtaposed internal 3ʹ and 5ʹ splice sites. In the first step, 92093, USA. -

(Bsc Zoology and Microbiology) Concept of Introns and Exons
Unit-5 Molecular Biology (BSc Zoology and Microbiology) Concept of introns and exons Most of the portion of a gene in higher eukaryotes consists of noncoding DNA that interrupts the relatively short segments of coding DNA. The coding sequences are called exons. The noncoding sequences are called introns. Intron: An intron is a portion of a gene that does not code for amino acids An intron is any nucleotide sequence within a gene which is represented in the primary transcript of the gene, but not present in the final processed form. In other words, Introns are noncoding regions of an RNA transcript which are eliminated by splicing before translation. Sequences that are joined together in the final mature RNA after RNA splicing are exons. Introns are very large chunks of RNA within a messenger RNA molecule that interfere with the code of the exons. And these introns get removed from the RNA molecule to leave a string of exons attached to each other so that the appropriate amino acids can be encoded for. Introns are rare in genes of prokaryotes. #Look carefully at the diagram above, we have already discussed about the modification and processing of eukaryotic RNA. In which 5’ guanine cap and 3’poly A tail is added. So at that time, noncoding regions i.e. introns are removed. We hv done ths already. Ok Exon: The coding sequences are called Exon. An exon is the portion of a gene that codes for amino acids. In the cells of plants and animals, most gene sequences are broken up by one or more DNA sequences called introns. -

Current Perspectives in Intronic Micro Rnas (Mirnas)
Journal of Biomedical Science (2006) 13:5–15 5 DOI 10.1007/s11373-005-9036-8 Current perspectives in intronic micro RNAs (miRNAs) Shao-Yao Ying & Shi-Lung Lin Department of Cell & Neurobiology, Keck School of Medicine, BMT-403, University of Southern California, 1333 San Pablo Street, Los Angeles, CA, 90033, USA Received 27 May 2005; accepted 14 September 2005 Ó 2005 National Science Council, Taipei Key words: fine-tuning of gene function, functional/structural genomics, gene expression, genetic regula- tion, intronic microRNA, miRNA biogenesis, miRNA, post-translational modification, regulatory gene Summary MicroRNAs (miRNAs), small single-stranded regulatory RNAs capable of interfering with intracellular messenger RNAs (mRNAs) that contain either complete or partial complementarity, are useful for the design of new therapies against cancer polymorphism and viral mutation. Numerous miRNAs have been reported to induce RNA interference (RNAi), a post-transcriptional gene silencing mechanism. Intronic miRNAs, derived from introns by RNA splicing and Dicer processing, can interfere with intracellular mRNAs to silence that gene expression. The intronic miRNAs differ uniquely from previously described intergenic miRNAs in the requirement of type II RNA polymerases (Pol-II) and spliceosomal components for its biogenesis. Several kinds of intronic miRNAs have been identified in Caenorhabditis elegans, mouse and human cells; however, neither their function nor application has been reported. To this day, the computer searching program for miRNA seldom include the intronic portion of protein-coding RNAs. The functional significance of artificially generated intronic miRNAs has been successfully ascertained in several biological systems such as zebrafishes, chicken embryos and adult mice, indicating the evolutionary pres- ervation of this gene regulation system in vivo. -

NSUN2 As the Methyltransferase and ALYREF As an M5c Reader
Cell Research (2017) 27:606-625. ORIGINAL ARTICLE www.nature.com/cr 5-methylcytosine promotes mRNA export — NSUN2 as the methyltransferase and ALYREF as an m5C reader Xin Yang1, 2, 3, *, Ying Yang2, *, Bao-Fa Sun2, *, Yu-Sheng Chen2, 3, *, Jia-Wei Xu1, 2, *, Wei-Yi Lai3, 4, *, Ang Li2, 3, Xing Wang2, 5, Devi Prasad Bhattarai2, 3, Wen Xiao2, Hui-Ying Sun2, Qin Zhu2, 3, Hai-Li Ma2, 3, Samir Adhikari2, Min Sun2, Ya-Juan Hao2, Bing Zhang2, Chun-Min Huang2, Niu Huang6, Gui-Bin Jiang4, Yong-Liang Zhao2, Hai-Lin Wang4, Ying-Pu Sun1, Yun-Gui Yang2, 3 1Center for Reproductive Medicine, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan 450000, China; 2Key Laboratory of Genomic and Precision Medicine, Collaborative Innovation Center of Genetics and Development, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; 3School of Life Science, University of Chinese Academy of Sciences, Beijing 100049, China; 4State Key Laboratory of Environmental Chemistry and Ecotoxicology, Research Center for Eco-Environmental Sciences, Chinese Academy of Sciences, Beijing 100085, China; 5Sino-Danish College, University of Chinese Academy of Sciences, Beijing 100049, China; 6National Institute of Biological Sciences, Beijing 102206, China 5-methylcytosine (m5C) is a post-transcriptional RNA modification identified in both stable and highly abundant tRNAs and rRNAs, and in mRNAs. However, its regulatory role in mRNA metabolism is still largely unknown. Here, we reveal that m5C modification is enriched in CG-rich regions and in regions immediately downstream of trans- lation initiation sites and has conserved, tissue-specific and dynamic features across mammalian transcriptomes. -

RNA Splice Sites Classification Using Convolutional Neural Network Models
RNA Splice Sites Classification Using Convolutional Neural Network Models Thanyathorn Thanapattheerakul Worrawat Engchuan Daniele Merico School of Information Technology, The Centre for Applied Genomics, Molecular Diagnostics, King Mongkut’s University of Genetics and Genome Biology, The Deep Genomics, Technology Thonburi, Hospital for Sick Children, Toronto, Ontario, Canada Bangkok, Thailand Toronto, Ontario, Canada [email protected] [email protected] [email protected] Narumol Doungpan Kiyota Hashimoto Jonathan H. Chan Faculty of Engineering, Faculty of Technology and School of Information Technology, King Mongkut’s University of Environment, King Mongkut’s University of Technology Thonburi, Prince of Songkla University, Technology Thonburi, Bangkok, Thailand Phuket, Thailand Bangkok, Thailand [email protected] [email protected] [email protected] Abstract—RNA splicing refers to the elimination of non- completely make it loss of function. The alternative splicing coding region on transcribed pre-messenger ribonucleic acid can produce different functional proteins, which could lead (RNA). Identifying splicing site is an essential step which can to causing abnormal states in human [3]. be used to gain novel insights of alternative splicing as well as Many studies have proposed models to recognize the splicing defects, potentially cause malfunction of protein splice sites to reveal which splice sites contain a mutation resulting from mutations at splice site. In this work, we that may cause a splicing error. One common method to propose a data preprocessing step applying to RNA sequences recognize binding sites in motif sequences is called Position- and the models leveraging Convolutional Neural Network (CNN). The preprocessing step includes reducing sequence Weight-Matrix (PWM). -

The CTD Role in Cotranscriptional RNA Processing and Surveillance
CORE Metadata, citation and similar papers at core.ac.uk Provided by Elsevier - Publisher Connector FEBS Letters 582 (2008) 1971–1976 Minireview The CTD role in cotranscriptional RNA processing and surveillance Se´rgio F. de Almeida, Maria Carmo-Fonseca* Instituto de Medicina Molecular, Faculdade de Medicina, Universidade de Lisboa, 1649-028 Lisboa, Portugal Received 4 April 2008; revised 13 April 2008; accepted 14 April 2008 Available online 22 April 2008 Edited by Ulrike Kutay proteins specific to each snRNP. The selection of specific splice Abstract In higher eukaryotes, the production of mature mes- senger RNA that exits the nucleus to be translated into protein sites (ss) on a particular pre-mRNA substrate relies on an intri- requires precise and extensive processing of the nascent tran- cate interplay involving the cooperative binding of trans-acting script. The processing steps include 50-end capping, splicing, splicing proteins to cis-acting sequence elements in the pre- and 30-end formation. Pre-mRNA processing is coupled to tran- mRNA. In mammals, an intron is defined by four short and scription by mechanisms that are not well understood but involve poorly conserved consensus sequences: the exon–intron junc- the carboxyl-terminal domain (CTD) of the largest subunit of tions (50ss and 30ss); the branch point sequence; and the poly- RNA polymerase II. This review focuses on recent findings that pyrimidine tract (Table 1). These sequences are recognized by provide novel insight into the role of the CTD in promoting RNA base pairing with the spliceosomal snRNAs (Fig. 1). In addi- processing and surveillance. tion, both exons and introns contain weak binding sites for a Ó 2008 Federation of European Biochemical Societies. -

Co-Transcriptional Loading of RNA Export Factors Shapes the Human Transcriptome
bioRxiv preprint doi: https://doi.org/10.1101/318709; this version posted May 10, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Title: Co-transcriptional loading of RNA export factors shapes the human transcriptome. Authors: Nicolas Viphakone1,$,*, Ian SudBery1,$, Catherine G. Heath1, David Sims2, Stuart A. Wilson1,*,# 1 Sheffield Institute For Nucleic Acids (SInFoNiA) and Department of Molecular Biology and Biotechnology, The University of Sheffield, Firth Court, Western Bank, Sheffield, UK. 2 MRC Computational Genomics Analysis and Training Programme (CGAT), MRC Centre for Computational Biology, MRC Weatherall Institute of Molecular Medicine, John Radcliffe Hospital, Headington, Oxford, OX3 9DS. $ Co-first author * Correspondence: [email protected], [email protected] # Lead contact Orcid Ids: N. Viphakone - 0000-0001-8219-837X, S. A. Wilson - 0000-0003-3073-258X, I. SudBery, 0000-0002-5038-0190 Key words: Nucleo-cytoplasmic transport, TREX, EJC, co-transcriptional splicing, alternative polyadenylation. 1 bioRxiv preprint doi: https://doi.org/10.1101/318709; this version posted May 10, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. ABSTRACT During gene expression, RNA export factors are mainly known for driving nucleocytoplasmic transport. Whilst early studies suggested that the Exon Junction Complex provides a binding platform for them, suBsequent work proposed that they are only recruited By the Cap-Binding Complex to the 5’ end of RNAs, as part of TREX. -
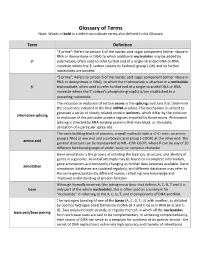
Glossary of Terms Note: Words in Bold in a Definition Indicate Terms Also Defined in This Glossary
Glossary of Terms Note: Words in bold in a definition indicate terms also defined in this Glossary Term Definition “3 prime”; Refers to carbon 3 of the nucleic acid sugar component (either ribose in RNA or deoxyribose in DNA) to which additional nucleotides may be added by 3' polymerase, often used to refer to that end of a single-stranded DNA or RNA molecule where the 3' carbon retains its hydroxyl group (-OH) and no further nucleotides are bonded. “5 prime”; Refers to carbon 5 of the nucleic acid sugar component (either ribose in RNA or deoxyribose in DNA), to which the triphosphate is attached in a nucleotide 5' triphosphate, often used to refer to that end of a single-stranded DNA or RNA molecule where the 5' carbon's phosphate group(s) is/are unattached to a preceding nucleotide. The inclusion or exclusion of certain exons in the splicing reactions that determine the sequences included in the final mRNA product. This mechanism is utilized to generate a series of closely related protein isoforms, which differ by the inclusion alternative splicing or exclusion of the particular protein regions encoded by those exons. Alternative splicing is directed by RNA-binding proteins that may block, or stimulate, utilization of a particular splice site. The basic building block of proteins, a small molecule with a -C-C- core, an amine group (-NH2) at one end and a carboxylic acid group (-COOH) at the other end. The amino acid general structure can be represented as NH2-CHR-COOH, where R can be any of 20 different functional groups of acidic, basic, or nonpolar character. -

Protein-Synthesis.Pdf
Protein Synthesis Contents Introduction ........................................................................................................................ 1 DNA Transcription ............................................................................................................... 1 The Primary Transcript ........................................................................................................ 4 Translation .......................................................................................................................... 7 Protein Structure............................................................................................................... 11 References ........................................................................................................................ 11 Resources .......................................................................................................................... 11 Introduction The genetic message carried on the DNA molecule is a code. This code is ultimately translated into a sequence of amino acids that, when complete, becomes a protein. Proteins carry out the “business” of the cell. Some proteins are used as structural components of cells, some are used to transport other molecules, still others are charged with directing chemical reactions. The latter class of proteins is the enzymes. Regardless of the role played by a protein in the cell one aspect is the same, they are all encoded in the base sequences of DNA. The path from DNA sequence to protein -

The Chloroplast Trans-Splicing RNA–Protein Supercomplex from the Green Alga Chlamydomonas Reinhardtii
cells Review The Chloroplast Trans-Splicing RNA–Protein Supercomplex from the Green Alga Chlamydomonas reinhardtii Ulrich Kück * and Olga Schmitt Allgemeine und Molekulare Botanik, Faculty for Biology and Biotechnology, Ruhr-Universität Bochum, Universitätsstraße 150, 44780 Bochum, Germany; [email protected] * Correspondence: [email protected]; Tel.: +49-234-32-28951 Abstract: In eukaryotes, RNA trans-splicing is a significant RNA modification process for the end-to- end ligation of exons from separately transcribed primary transcripts to generate mature mRNA. So far, three different categories of RNA trans-splicing have been found in organisms within a diverse range. Here, we review trans-splicing of discontinuous group II introns, which occurs in chloroplasts and mitochondria of lower eukaryotes and plants. We discuss the origin of intronic sequences and the evolutionary relationship between chloroplast ribonucleoprotein complexes and the nuclear spliceosome. Finally, we focus on the ribonucleoprotein supercomplex involved in trans-splicing of chloroplast group II introns from the green alga Chlamydomonas reinhardtii. This complex has been well characterized genetically and biochemically, resulting in a detailed picture of the chloroplast ribonucleoprotein supercomplex. This information contributes substantially to our understanding of the function of RNA-processing machineries and might provide a blueprint for other splicing complexes involved in trans- as well as cis-splicing of organellar intron RNAs. Keywords: group II intron; trans-splicing; ribonucleoprotein complex; chloroplast; Chlamydomonas reinhardtii Citation: Kück, U.; Schmitt, O. The Chloroplast Trans-Splicing RNA–Protein Supercomplex from the Green Alga Chlamydomonas reinhardtii. 1. Introduction Cells 2021, 10, 290. https://doi.org/ One of the unexpected and outstanding discoveries in 20th century biology was the 10.3390/cells10020290 identification of discontinuous eukaryotic genes [1,2]. -

Alternatively Spliced Genes
1 Alternatively Spliced Genes Jane Y. Wu1,2,LiyaYuan1 and Necat Havlioglu1 1Washington University School of Medicine, St. Louis, MO, USA 2John F. Kennedy Center for Research on Human Development, Vanderbilt University Medical Center, Nashville, TN, USA 1 Pre-mRNA Splicing and Splicing Machinery 3 1.1 Splicing Machinery: Spliceosome 3 1.2 Splicing Signals 4 1.3 Spliceosomal UsnRNP Biogenesis 12 1.4 Spliceosome Assembly 13 1.5 Biochemical Mechanisms of pre-mRNA Splicing 17 2 Alternative pre-mRNA Splicing 17 2.1 Alternative Splicing and its Role in Regulating Gene Activities and Generating Genetic Diversity 17 2.1.1 Different Patterns of Alternative Splicing 17 2.1.2 Alternative Splicing and Genetic Diversity 18 2.2 Mechanisms Underlying Alternative Splicing Regulation 19 2.2.1 Splicing Signals and Splicing Regulatory Elements 20 2.2.2 Trans-acting Splicing Regulators 23 2.3 Tissue-specific and Developmentally Regulated Alternative Splicing 26 2.4 Regulation of Alternative Splicing in Response to Extracellular Stimuli 27 3 Pre-mRNA Splicing and Human Diseases 28 3.1 Splicing Defects in Human Diseases 28 3.2 Molecular Mechanisms Underlying Splicing Defects Associated with Disease 33 4 Perspectives on Diagnosis and Treatment of Diseases Caused by pre-mRNA Splicing Defects 36 4.1 Diagnosis of Human Diseases Caused by Splicing Defects 36 2 Alternatively Spliced Genes 4.2 Potential Therapeutic Approaches 37 4.2.1 Oligonucleotide-based Approaches: Antisense, RNAi, and Chimeric Molecules 37 4.2.2 Ribozymes 37 4.2.3 SMaRT 38 4.2.4 Chemical Compounds 38 5 Concluding Remarks 38 Acknowledgment 39 Bibliography 39 Books and Reviews 39 Keywords Pre-mRNA Nascent transcripts that are precursors of mature messenger RNAs. -

Principles of RNA Processing from Analysis of Enhanced CLIP Maps for 150 RNA Binding Proteins Eric L
Van Nostrand et al. Genome Biology (2020) 21:90 https://doi.org/10.1186/s13059-020-01982-9 RESEARCH Open Access Principles of RNA processing from analysis of enhanced CLIP maps for 150 RNA binding proteins Eric L. Van Nostrand1,2, Gabriel A. Pratt1,2, Brian A. Yee1,2, Emily C. Wheeler1,2, Steven M. Blue1,2, Jasmine Mueller1,2, Samuel S. Park1,2, Keri E. Garcia1,2, Chelsea Gelboin-Burkhart1,2, Thai B. Nguyen1,2, Ines Rabano1,2, Rebecca Stanton1,2, Balaji Sundararaman1,2, Ruth Wang1,2, Xiang-Dong Fu1,2, Brenton R. Graveley3* and Gene W. Yeo1,2* Abstract Background: A critical step in uncovering rules of RNA processing is to study the in vivo regulatory networks of RNA binding proteins (RBPs). Crosslinking and immunoprecipitation (CLIP) methods enable mapping RBP targets transcriptome-wide, but methodological differences present challenges to large-scale analysis across datasets. The development of enhanced CLIP (eCLIP) enabled the mapping of targets for 150 RBPs in K562 and HepG2, creating a unique resource of RBP interactomes profiled with a standardized methodology in the same cell types. Results: Our analysis of 223 eCLIP datasets reveals a range of binding modalities, including highly resolved positioning around splicing signals and mRNA untranslated regions that associate with distinct RBP functions. Quantification of enrichment for repetitive and abundant multicopy elements reveals 70% of RBPs have enrichment for non-mRNA element classes, enables identification of novel ribosomal RNA processing factors and sites, and suggests that association with retrotransposable elements reflects multiple RBP mechanisms of action. Analysis of spliceosomal RBPs indicates that eCLIP resolves AQR association after intronic lariat formation, enabling identification of branch points with single-nucleotide resolution, and provides genome-wide validation for a branch point-based scanning model for 3′ splice site recognition.