Using Edna to Biomonitor the Fish Community in a Tropical Oligotrophic Lake
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

CAT Vertebradosgt CDC CECON USAC 2019
Catálogo de Autoridades Taxonómicas de vertebrados de Guatemala CDC-CECON-USAC 2019 Centro de Datos para la Conservación (CDC) Centro de Estudios Conservacionistas (Cecon) Facultad de Ciencias Químicas y Farmacia Universidad de San Carlos de Guatemala Este documento fue elaborado por el Centro de Datos para la Conservación (CDC) del Centro de Estudios Conservacionistas (Cecon) de la Facultad de Ciencias Químicas y Farmacia de la Universidad de San Carlos de Guatemala. Guatemala, 2019 Textos y edición: Manolo J. García. Zoólogo CDC Primera edición, 2019 Centro de Estudios Conservacionistas (Cecon) de la Facultad de Ciencias Químicas y Farmacia de la Universidad de San Carlos de Guatemala ISBN: 978-9929-570-19-1 Cita sugerida: Centro de Estudios Conservacionistas [Cecon]. (2019). Catálogo de autoridades taxonómicas de vertebrados de Guatemala (Documento técnico). Guatemala: Centro de Datos para la Conservación [CDC], Centro de Estudios Conservacionistas [Cecon], Facultad de Ciencias Químicas y Farmacia, Universidad de San Carlos de Guatemala [Usac]. Índice 1. Presentación ............................................................................................ 4 2. Directrices generales para uso del CAT .............................................. 5 2.1 El grupo objetivo ..................................................................... 5 2.2 Categorías taxonómicas ......................................................... 5 2.3 Nombre de autoridades .......................................................... 5 2.4 Estatus taxonómico -

Participación Comunitaria En La Transferencia Tecnológica De Un Sistema Acuícola De Peces JAINA Costas Y Mares Ante El Cambio Climático, ÁTICO Nativos
JAINA Costas y Mares ante el Cambio Climático 2(1), 2020 López Jiménez, L.N., Maldonado Romo, A., Álvarez-González, C.A., Peña Marín, E.S., Fernández-Montes de Oca, A., 2020. Participación comunitaria en la transferencia tecnológica de un sistema acuícola de peces JAINA Costas y Mares ante el Cambio Climático, ÁTICO nativos. 2(1): 31-46. JAINACOSTAS Y MARES ANTE EL CAMBIO CLIM doi 10.26359/52462.0320 Reporte de Investigación / Research Report Participación comunitaria en la transferencia tecnológica de un sistema acuícola de peces nativos Community involvement in technological transference ofLeonardo a native Noriel López fish Jiménez aquaculture1, Axel Maldonado Romo 2system, Carlos Alfonso Álvarez-González3, Emyr Saúl Peña Marín3,4 y Ana Fernández-Montes de Oca5 1 Centro del Cambio Global y la Sustentabilidad, A.C. 2 Universidad Autónoma Chapingo. 3 Laboratorio de Acuicultura Tropical, DACBIOL, Universidad Juárez Autónoma de Tabasco. 4 Cátedra CONACyT- Laboratorio de Acuacultura, DACBIOL, Universidad Juárez Autónoma de Tabasco. 5 Laboratorio de Sistemas de Información Geográfica. Instituto de Biología, UNAM. * autor de correspondencia: [email protected] doi 10.26359/52462.0320 Recibido 15/enero/2020. Aceptado 14/julio/2020 JAINA Costas y Mares ante el Cambio Climático Coordinación editorial de este número: Edgar Mendoza Franco Este es un artículo bajo licencia Creative Commons CC BY-NC-ND. 31 JAINA costas y mares ante el cambio climático 2(1): 31-46 López Jiménez et al. Resumen La acuacultura de baja escala puede ser un modelo exitoso en ambientes rurales porque puede atenuar la deman- da de productos y la generación de empleos, pero uno de los factores limitantes para establecer estos sistemas de acuicultura es la debilidad sobre el manejo de los procesos tecnológicos para la producción de peces por parte de los productores. -

They're Not Just Convicts Anymore
2008 FAAS Publication Awards. Please follow reprint instructions at http://www.faas.info/2008_publication_awards_winners.html#reprintpolicy They’re Not Just Convicts Anymore By Daniel Spielman All Photos by Daniel Spielman Introduction Convicts get no respect, with many a cichlidophile turning up his or her nose at the sight of a group on Convicts in a tank or a bag of fry in an auction. It’s time for that to change. Easy to breed and exhibiting wonderful parental care, Convict cichlids (Cryptoheros nigrofasciatus) have long been staples in the hobby. Indeed, for many new fishkeepers, the satisfaction of watching a pair of Convict parents herd a group of fry around a tank sparks the initial desire to keep other mem bers of the cichlid family. Indeed, my fish cichlids were Convicts, and watch ing them care for their fry definitely got me hooked. The contrast with trying to keep guppies or swordtails from eating their own offspring is striking (how did eating one’s own young ever evolve in the first place?). However, the very fact that Convicts spawn so readily in the aquarium causes many hobbyists to quickly lose interest in the species. Although Convicts occasionally appear on experienced hobbyists’ lists of most favorite cichlid, the problem is they are just too common and too easy to breed. Typical Convict spawning jokes involve two fish and a wet paper towel, and one often feels fortunate to break the $1 barrier at the auction to avoid the indignity of having to bring one’s fish back home again at the end of the monthly club meeting. -

Hotspots, Extinction Risk and Conservation Priorities of Greater Caribbean and Gulf of Mexico Marine Bony Shorefishes
Old Dominion University ODU Digital Commons Biological Sciences Theses & Dissertations Biological Sciences Summer 2016 Hotspots, Extinction Risk and Conservation Priorities of Greater Caribbean and Gulf of Mexico Marine Bony Shorefishes Christi Linardich Old Dominion University, [email protected] Follow this and additional works at: https://digitalcommons.odu.edu/biology_etds Part of the Biodiversity Commons, Biology Commons, Environmental Health and Protection Commons, and the Marine Biology Commons Recommended Citation Linardich, Christi. "Hotspots, Extinction Risk and Conservation Priorities of Greater Caribbean and Gulf of Mexico Marine Bony Shorefishes" (2016). Master of Science (MS), Thesis, Biological Sciences, Old Dominion University, DOI: 10.25777/hydh-jp82 https://digitalcommons.odu.edu/biology_etds/13 This Thesis is brought to you for free and open access by the Biological Sciences at ODU Digital Commons. It has been accepted for inclusion in Biological Sciences Theses & Dissertations by an authorized administrator of ODU Digital Commons. For more information, please contact [email protected]. HOTSPOTS, EXTINCTION RISK AND CONSERVATION PRIORITIES OF GREATER CARIBBEAN AND GULF OF MEXICO MARINE BONY SHOREFISHES by Christi Linardich B.A. December 2006, Florida Gulf Coast University A Thesis Submitted to the Faculty of Old Dominion University in Partial Fulfillment of the Requirements for the Degree of MASTER OF SCIENCE BIOLOGY OLD DOMINION UNIVERSITY August 2016 Approved by: Kent E. Carpenter (Advisor) Beth Polidoro (Member) Holly Gaff (Member) ABSTRACT HOTSPOTS, EXTINCTION RISK AND CONSERVATION PRIORITIES OF GREATER CARIBBEAN AND GULF OF MEXICO MARINE BONY SHOREFISHES Christi Linardich Old Dominion University, 2016 Advisor: Dr. Kent E. Carpenter Understanding the status of species is important for allocation of resources to redress biodiversity loss. -

Patterns of Evolution in Gobies (Teleostei: Gobiidae): a Multi-Scale Phylogenetic Investigation
PATTERNS OF EVOLUTION IN GOBIES (TELEOSTEI: GOBIIDAE): A MULTI-SCALE PHYLOGENETIC INVESTIGATION A Dissertation by LUKE MICHAEL TORNABENE BS, Hofstra University, 2007 MS, Texas A&M University-Corpus Christi, 2010 Submitted in Partial Fulfillment of the Requirements for the Degree of DOCTOR OF PHILOSOPHY in MARINE BIOLOGY Texas A&M University-Corpus Christi Corpus Christi, Texas December 2014 © Luke Michael Tornabene All Rights Reserved December 2014 PATTERNS OF EVOLUTION IN GOBIES (TELEOSTEI: GOBIIDAE): A MULTI-SCALE PHYLOGENETIC INVESTIGATION A Dissertation by LUKE MICHAEL TORNABENE This dissertation meets the standards for scope and quality of Texas A&M University-Corpus Christi and is hereby approved. Frank L. Pezold, PhD Chris Bird, PhD Chair Committee Member Kevin W. Conway, PhD James D. Hogan, PhD Committee Member Committee Member Lea-Der Chen, PhD Graduate Faculty Representative December 2014 ABSTRACT The family of fishes commonly known as gobies (Teleostei: Gobiidae) is one of the most diverse lineages of vertebrates in the world. With more than 1700 species of gobies spread among more than 200 genera, gobies are the most species-rich family of marine fishes. Gobies can be found in nearly every aquatic habitat on earth, and are often the most diverse and numerically abundant fishes in tropical and subtropical habitats, especially coral reefs. Their remarkable taxonomic, morphological and ecological diversity make them an ideal model group for studying the processes driving taxonomic and phenotypic diversification in aquatic vertebrates. Unfortunately the phylogenetic relationships of many groups of gobies are poorly resolved, obscuring our understanding of the evolution of their ecological diversity. This dissertation is a multi-scale phylogenetic study that aims to clarify phylogenetic relationships across the Gobiidae and demonstrate the utility of this family for studies of macroevolution and speciation at multiple evolutionary timescales. -

Changes in the Fish Community of a Western Caribbean Estuary After the Expansion of an Artificial Channel to the Sea
water Article Changes in the Fish Community of a Western Caribbean Estuary after the Expansion of an Artificial Channel to the Sea Juan J. Schmitter-Soto * and Roberto L. Herrera-Pavón El Colegio de la Frontera Sur, Av. Centenario km 5.5, Chetumal 77014, Quintana Roo, Mexico; [email protected] * Correspondence: [email protected]; Tel.: +52-983-835-0440 (ext. 4302) Received: 30 October 2019; Accepted: 2 December 2019; Published: 6 December 2019 Abstract: Increased connectivity between coastal lagoons and the sea is expected to entail a greater proportion of marine species in the former. Chetumal Bay, estuary of the Hondo river into the Caribbean, had a limited access to the sea until the opening of the Zaragoza Canal. We sought changes in the fish community from 1999–2001 (just after an expansion of the canal) to 2015–2018. The same fishing gear was used, in the same localities, during all seasons. Total fish abundance and mean local richness decreased, although total abundance increased in the polyhaline zone. Diversity was greater in the oligohaline zone in 1999–2001, and in the mesohaline zone in 2015–2018. Three guilds were absent in 2015–2018: Medium-sized herbivores, large piscivores, and medium-sized planktivores. Abundance of small benthivores decreased by decade; medium-sized piscivores and small planktivores became more abundant in 2015–2018 in the polyhaline zone. These changes may be due to the opening of the channel, but illegal fishing outside the bay may explain the decrease in juveniles of large piscivores, and erosion in the innermost part may be destroying important habitats. -

2019-JMIH-Program-Book-MASTER
W:\CNCP\People\Richardson\FY19\JMIH - Rochester NY\Program\2018 JMIH Program Book.pub 2 Organizing Societies American Elasmobranch Society 34th Annual Meeting President: Dave Ebert Treasurer: Christine Bedore Secretary: Tonya Wiley Editor and Webmaster: Chuck Bangley Immediate Past President: Dean Grubbs American Society of Ichthyologists and Herpetologists 98th Annual Meeting President: Kathleen Cole President Elect: Chris Beachy Past President: Brian Crother Prior Past President: Carole Baldwin Treasurer: Katherine Maslenikov Secretary: Prosanta Chakrabarty Editor: W. Leo Smith Herpetologists’ League 76th Annual Meeting President: Willem Roosenburg Vice-President: Susan Walls Immediate Past President: David Sever (deceased) Secretary: Renata Platenburg Treasurer: Laurie Mauger Communications Secretary: Max Lambert Herpetologica Editor: Stephen Mullin Herpetological Monographs Editor: Michael Harvey Society for the Study of Amphibians and Reptiles 61th Annual Meeting President: Marty Crump President-Elect: Kirsten Nicholson Immediate Past-President: Richard Shine Secretary: Marion R. Preest Treasurer: Ann V. Paterson Publications Secretary: Cari-Ann Hickerson 3 Thanks to our Sponsors! PARTNER SPONSOR SUPPORTER SPONSOR 4 We would like to thank the following: Local Hosts Alan Savitzky, Utah State University, LHC Co-Chair Catherine Malone, Utah State University, LHC Co-Chair Diana Marques, Local Host Logo Artist Marty Crump, Utah State University Volunteers We wish to thank the following volunteers who have helped make the Joint Meeting -

Suborder GOBIOIDEI ELEOTRIDAE Sleepers by E.O
click for previous page 1778 Bony Fishes Suborder GOBIOIDEI ELEOTRIDAE Sleepers by E.O. Murdy, National Science Foundation, Virginia, USA and D.F. Hoese, Australian Museum, Sydney, Australia iagnostic characters: Small to medium-sized (most do not exceed 20 cm, although Gobiomorus from Dthis area may reach 60 cm). Typically, body stout; head short and broad; snout blunt; gill membranes broadly joined to isthmus. Teeth usually small, conical and in several rows in jaws. Six branchiostegal rays. Two separate dorsal fins, first dorsal fin with 6 or 7 weak spines, second dorsal fin with 1 weak spine followed by 6 to 12 soft rays; second dorsal fin and anal fin relatively short-based; origin of anal fin just posterior to vertical with origin of second dorsal fin; terminal ray of second dorsal and anal fins divided to its base (but counted as a single element);anal fin with 1 weak spine followed by 6 to 12 soft rays;caudal fin broad and rounded, compris- ing 15 or 17 segmented rays; pectoral fin broad with 14 to 25 soft rays; pelvic fin long with 1 spine and 5 soft rays.Pelvic fins separate and not connected by a membrane.Scales large and either cycloid or ctenoid.No lateral line on body. Head typically scaled, scales being either cycloid or ctenoid with a series of sensory ca- nals and pores as well as cutaneous papillae. Colour: not brightly coloured, most are light or dark brown or olive with some metallic glints. Habitat, biology, and fisheries: Typically occur in fresh or brackish waters, although some species are truly marine. -

Genetic Variations of Lansium Domesticum Corr
BIODIVERSITAS ISSN: 1412-033X Volume 19, Number 6, November 2018 E-ISSN: 2085-4722 Pages: 2252-2274 DOI: 10.13057/biodiv/d190634 Ichthyofauna checklist (Chordata: Actinopterygii) for indicating water quality in Kampar River catchment, Malaysia CASEY KEAT-CHUAN NG♥, PETER AUN-CHUAN OOI, WEY-LIM WONG, GIDEON KHOO♥♥ Faculty of Science, Universiti Tunku Abdul Rahman. Jl. Universiti Bandar Barat, 31900 Kampar, Perak, Malaysia. Tel.: +605-4688888, Fax.: +605-4661313. ♥email: [email protected], ♥♥ [email protected] Manuscript received: 18 August 2018. Revision accepted: 12 November 2018. Abstract. Ng CKC, Ooi PAC, Wong WL, Khoo G. 2018. Ichthyofauna checklist (Chordata: Actinopterygii) for indicating water quality in Kampar River catchment, Malaysia. Biodiversitas 19: 2252-2274. The limnological habitats are receptors of pollution, thus local fish species richness is a plausible biological indicator to reflect the quality of a particular water body. However, database on species occurrence that corresponds with the water physico-chemistry constituents is often not available. The problem is compounded by the lack of species identification description to assist those working on river and freshwater resource conservation projects. This paper attempts to fill the gaps in the context of Kampar River drainage. Based on sampling exercises conducted from October 2015 to March 2017, an annotated list with visual data for 56 species belonging to 44 genera and 23 families is presented. The water physico-chemistry data is also summarized with the corresponding visual data of limnological zones studied. The species diversity results are further compared with other local drainages and the correlation between area size and their relationship is expressed by y = 17.627e0.0601x. -

Neue Gattungseinteilung Der Mittelamerikanischen Cichliden
DCG_Info_07_2016_HR_20160621_DCG_Info 21.06.2016 06:51 Seite 146 Neue Gattungseinteilung der mittelamerikanischen Cichliden Rico Morgenstern Foto: Juan Miguel Artigas Azas Theraps irregularis verbleibt als einzige Art in der Gattung Theraps. Die Aufnahme entstand im Rio Lacanja im südlichen Chiapas, Mexiko. Inzwischen ist es fast 33 Jahre her, wenigstens Versuche, einzelne Gat- schien die Abhandlung „Diversity and dass KULLANDER (1983) die Gattung tungen neu zu definieren – aber eine evolution of the Middle American cich- Cichlasoma auf zwölf südamerikani- umfassende Gesamtbearbeitung er- lid fishes (Teleostei: Cichlidae) with re- sche, nahe mit der Typusart C. bima- folgte bisher nicht. Vielfach wurden vised classification“ (ŘIČAN et al. culatum verwandte Arten beschränkte. Zuordnungen vorgenommen, ohne 2016). Seither durfte der Name streng ge- dass man sich um eine wirkliche Be- nommen für die Mehrzahl der bis gründung bemühte. Die Arbeit berücksichtigt alle Cichliden dahin in dieser ehemaligen Sammel- Nord- und Mittelamerikas und der An- gattung untergebrachten, überwie- Bei der Gattungseinteilung der mittel- tillen sowie einige eng verwandte süd- gend mittelamerikanischen Arten amerikanischen Cichliden herrschte amerikanische Gattungen (Australoheros, nicht mehr verwendet werden. Man- somit bis vor kurzem ein ziemliches Caquetaia, Heroina, Mesoheros). gels geeigneter Alternativen ist das Chaos. Nun ist jedoch das Ende der An- Diese Fische gehören zu den „heroinen aber dennoch geschehen, wobei der führungszeichen (mit einer Ausnahme) -

View/Download
CICHLIFORMES: Cichlidae (part 6) · 1 The ETYFish Project © Christopher Scharpf and Kenneth J. Lazara COMMENTS: v. 6.0 - 18 April 2020 Order CICHLIFORMES (part 6 of 8) Family CICHLIDAE Cichlids (part 6 of 7) Subfamily Cichlinae American Cichlids (Acarichthys through Cryptoheros) Acarichthys Eigenmann 1912 Acara (=Astronotus, from acará, Tupí-Guaraní word for cichlids), original genus of A. heckelii; ichthys, fish Acarichthys heckelii (Müller & Troschel 1849) in honor of Austrian ichthyologist Johann Jakob Heckel (1790-1857), who proposed the original genus, Acara (=Astronotus) in 1840, and was the first to seriously study cichlids and revise the family Acaronia Myers 1940 -ia, belonging to: Acara (=Astronotus, from acará, Tupí-Guaraní word for cichlids), original genus of A. nassa [replacement name for Acaropsis Steindachner 1875, preoccupied by Acaropsis Moquin-Tandon 1863 in Arachnida] Acaronia nassa (Heckel 1840) wicker basket or fish trap, presumably based on its local name, Bocca de Juquia, meaning “fish trap mouth,” referring to its protractile jaws and gape-and-suck feeding strategy Acaronia vultuosa Kullander 1989 full of facial expressions or grimaces, referring to diagnostic conspicuous black markings on head Aequidens Eigenmann & Bray 1894 aequus, same or equal; dens, teeth, referring to even-sized teeth of A. tetramerus, proposed as a subgenus of Astronotus, which has enlarged anterior teeth Aequidens chimantanus Inger 1956 -anus, belonging to: Chimantá-tepui, Venezuela, where type locality (Río Abácapa, elevation 396 m) is -
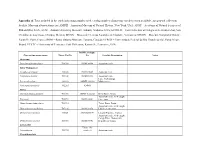
Appendix A. Taxa Included in the Study Indicating Samples Used, Catalog Number of Museum Vouchers When Available, and General Collection Locality
Appendix A. Taxa included in the study indicating samples used, catalog number of museum vouchers when available, and general collection locality. Museum abbreviations are: AMNH – American Museum of Natural History, New York, USA; ANSP – Academy of Natural Sciences of Philadelphia, USA; AUM – Auburn University Museum, Auburn, Alabama, USA; ECOSUR – Fish Collection at Colegio de la Frontera Sur, San Cristóbal de Las Casas, Chiapas, Mexico; MCNG – Museo de Ciencias Naturales de Guanare, Venezuela; MNHN – Muséum National d’Histoire Naturelle, Paris, France; ROM – Royal Ontario Museum, Toronto, Canada; UFRGS – Universidade Federal do Rio Grande do Sul, Porto Alegre, Brazil; UTFTC – University of Tennessee Fish Collection, Knoxville, Tennessee, USA. ROM Catalogue Current taxonomy name Tissue Cat No No Locality Description Notes Outgroup Pseudetroplus maculatus T14743 ROM 98998 Aquarium trade India-Madagascar Etroplus suratensis T13505 ROM 93809 Aquarium trade Paratilapia polleni T13100 ROM 88333 Aquarium trade Lake Andrapongy, Paretroplus damii 201936 AMNH 201936 Madagascar Paretroplus polyactis T12265 AMNH Africa Chromidotilapia guntheri T11700 AMNH I-226361 Beffa River, Benin Aquarium trade, wild caught, Etia nguti T10792 ROM 88042 Cameroon Hemichromis bimaculatus T11719 Tchan Duga, Benin Aquarium trade, wild caught, Heterochromis multidens T07136 ROM 88350 Lobeke, Cameroon Oreochromis niloticus 9092S AMNH254194 Littoral Province, Guinea Aquarium trade, wild caught, Congo River, Democratic Orthochromis stormsi T10766 ROM 88041 Republic of