Report Cover
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Department of Education and Science of Ukraine National University of Food Technologies
DEPARTMENT OF EDUCATION AND SCIENCE OF UKRAINE NATIONAL UNIVERSITY OF FOOD TECHNOLOGIES APPROVED _______________A.I. Ukrainets (signature) «_____» ___________2016 y. V.N. ISCHENKO М.S. SОVКО N.V. ISCHENKO EQUIPMENT AND SAFETY IN CHEMICAL LABORATORY All quotations, digital and actual material, bibliographical information are checked. Writing units respond to the standards Signature (s) of the author (s) _____________________ « _17 » __May__2016 y. Registration number of electronic textbook in EMM 59. 04 – 07. 06. 2016 KIEV NUFT 2016 UDC 542.07 Recommended by the Academic Council of the National University of Food Technologies as learner’s guide for students of higher educational establishments (Minutes № 13 from 31.05. 2016) Reviewers: V.I Maksin – Dr. of Chemistry, Professor of the Department of Bioinorganic Chemistry and Water Quality NUBaN Ukraine O.O. Tkachuck – Ph.D. in Chemistry, the head of the laboratory ofnational anti-doping center of Ukraine O.V. Berezovska – senior lecturer of the Department of Business Foreign Language and International Communication NUFT IschenkoV.N. Equipment and safety in chemical laboratory [Electronic resourse]:learner’s guide /V.N. Ischenko, М.S. Sоvко, N.V. Ischenko. – K.: NUFT, 2016. –27 р. The learner’s guide provides information about modern chemical laboratory equipment. Chemical laboratory ware and equipment necessary for conducting laboratory work are described. The guide provides the rules of behavior and safety in chemical laboratory, rules of work with chemicals Learner’s guide is recommended for students of academic level of bachelor, who study Chemistry in English. Authors: V.N. ISCHENKO, PhD М.S. SОVКО, N.V. ISCHENKO, PhD Introduced in author’s edition © V.N. -

Food Protection Trends 2010-09: Vol 30 Iss 9
September 2010 ee Vol. a 30, No. 9 Periodicals 6200 Aurora Avenue ¢ Suite 200W Des Moines, lowa 50322-2864, USA merle Mm manele drele Mm igsalels Science and NewS trom the international Association for Food Protection Inter-agency Public Health Collaboration Pathogens Used with Biltong Product www.foodprotection.org BD |; Microbiology Media Solutions for Food Safety Reduce the risk to consumer safety with BBL Campy- e Over 170 years of combined Difco” and Cefex Agar* prepared plated medium for the isolation, BBL” microbiology experience enumeration and detection of Camplyobacter species directly from poultry. ' Microbiology— It’s what we do e Adopted by the National Advisory Committee on Microbiological Criteria for Foods for the isolation Find out what we can do for you. of Campylobacter species from chicken carcasses! Visit us on the web at www.bd.com/ds ¢ Better selectivity and easier differentiation of C. jejuni from other flora? BD Diagnostics 800.638.8663 *U.S. Patent No. 5,891,709 licensed to Becton, Dickinson and Company www.bd.com/ds 1 aa.) OAS A Food Process Control Microorganisms When the world’s leading food manufacturers need to meet expanding regulatory, liability, and quality assurance challenges they put their trust in Microbiologics. Our food process controls are guaranteed to deliver a specific number of colony forming units. Each easy-to-use microorganism preparation is authentic and traceable to an original reference strain. EZ-FPC™ * Qualitative preparations for Presence/Absence Testing * Quantitative preparations -

Seed-Bag Experiment Arnold Lab, University of Arizona Contact Arnold
Symbiosis in the Context of an Invasive, Non-Native Grass: Fungal Biodiversity and Student Engagement Item Type text; Electronic Thesis Authors Lehr, Gavin Charles Publisher The University of Arizona. Rights Copyright © is held by the author. Digital access to this material is made possible by the University Libraries, University of Arizona. Further transmission, reproduction or presentation (such as public display or performance) of protected items is prohibited except with permission of the author. Download date 26/09/2021 11:52:51 Link to Item http://hdl.handle.net/10150/626728 Seed-bag experiment Arnold Lab, University of Arizona Contact [email protected] before use in publication This version: Ming-min Lee and Betsy Arnold, November 2016 Goals of this project: To collect seed-associated microbes from arid agricultural and unimproved soils in Arizona; to catalog these microbes, to determine their effects on seed germination, and to analyze site metadata to discover any patterns in positive and negative microbe-seed associations. I. Seed Bag Construction Purpose: To construct sterile seed bags for seed burial. Bags allow soil and microbes to contact seeds while excluding insects and other arthropods that might damage seeds. Number of bags: 18 bags per plot are required, along with two bags to be used as field controls and an additional two as lab controls. We typically have 2 or 3 plots per site. Materials: 50μm polyester filter material (https://www.dudadiesel.com/search.php?query=%2Bpolyester+%2Bsheet) Scissors Rulers Staplers and staples Aluminum foil Laminar flow hood or alcohol burner Forceps, spoonulas, teaspoons etc. Infrared or bead sterilizer, or alcohol burner and ETOH for sterilizing tools Autoclave Seeds 1. -

CHEM C3000 Manual
EXPERIMENT MANUAL Version 2.0 Please observe the safety information, the advice for supervising adults on page 5, the safety rules on page 6, the information about hazardous substances and mixtures (chemicals) on pages 7-9 and their environmentally sound disposal on pages 175-177, the safety for experiments with batteries on page 192, the first aid information on the inside front cover and the instructions on the use of the alcohol burner on page 12. WARNING. Not suitable for children under 12 years. For use under adult supervision. Contains some chemicals which present a hazard to health. Read the instructions before use, follow them and keep them for reference. Do not allow chemicals to come into contact with any part of the body, particularly the mouth and eyes. Keep small children and animals away from experiments. Keep the experimental set out of the reach of children under 12 years old. Eye protection for supervising adults is not included. WARNING — Chemistry Set. This set contains chemicals and parts that may be harmful if misused. Read cautions on individual containers and in manual carefully. Not to be used by children except under adult supervision. Franckh-Kosmos Verlags-GmbH & Co. KG, Pfizerstr. 5-7, 70184 Stuttgart, Germany | +49 (0) 711 2191-0 | www.kosmos.de Thames & Kosmos, 301 Friendship St., Providence, RI, 02903, USA | 1-800-587-2872 | www.thamesandkosmos.com Thames & Kosmos UK Ltd, Goudhurst, Kent, TN17 2QZ , United Kingdom | 01580 212000 | www.thamesandkosmos.co.uk Contents Safety and Information First Aid Information . Inside front cover Poison Control Contact Information . Inside front cover Advice for Supervising Adults . -

JOSEPH WANJALA KILWAKE THESIS.Pdf
i ASSESSMENT OF WATER QUALITY IN BOREHOLES AND WELLS IN WAA LOCATION, KWALE COUNTY - KENYA JOSEPH WANJALA KILWAKE A thesis submitted in partial fulfillment of the requirements for the Degree of Masters of Environmental Science of Pwani University May, 2016 ii iii DEDICATION This work is dedicated to my wife, Everlyne, son Ian and late grandfather Patroba for their love, sacrifice, prayers and support in my study. iv ACKNOWLEDGEMENTS I would like to acknowledge and thank my supervisors Prof. Mwakio Tole and Dr. Okeyo Benards for guidance, encouragement and support in my study period. I am indebted to Prof. Halimu Shauri and Dr. Maarifa Mwakumanya of Pwani University for the support in proposal development. I register my special gratitude to Mrs. Salome Mwaruwa, Principal Waa Girls' School for support and encouragement, Mr. Ali Harun, Hamisi Masito, Ali Mshindo and Juma Kanga during mapping, data collection and for linkage with the local community. I do appreciate Mr. Patrick Oduma, OCPD Kwale District, Dr. Remmy Shiundu, Barasa Joel, Twahir Mohammed, Eng. John Wanjala (EPHRAIM Company) and Josephat Orina for material support and advise. I gratefully thank Miss Nyambura Mwangi, Mr. Peter Karanja (Pwani University) and David Bett (Fisheries Department) for support in sample collection, data analysis and consultation. I sincerely thank the Almighty God for the guidance in my study. v ABSTRACT Water from boreholes and dug wells is extensively used in Kwale County, especially by rural communities living away from established market centers, where piped water is commonly available. The study aimed to assess the quality of water in boreholes and dug wells found in Waa location of Kwale County – Kenya. -
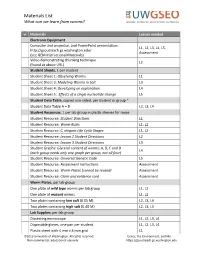
Materials List and Instructions for Maintaining Worm Stocks
Materials List What can we learn from worms? Materials Lesson needed Electronic Equipment Computer and projector, and PowerPoint presentation L1, L2, L3, L4, L5, http://gsoutreach.gs.washington.edu/ Assessment (see GEM Instructional Materials) Video demonstrating chunking technique L2 (found at above URL) Student Sheets, 1 per student Student Sheet 1: Observing Worms L1 Student Sheet 3: Modeling Worms in Salt L3 Student Sheet 4: Developing an explanation L4 Student Sheet 5: Effects of a single nucleotide change L5 Student Data Table, copied one-sided, per student or group * Student Data Table A – D L2, L3, L4 Student Resources, 1 per lab group in plastic sleeves for reuse Student Resource: Student Directions L1 Student Resource: Worm Rules L1, L2 Student Resource: C. elegans Life Cycle Stages L1, L2 Student Resource: Lesson 2 Student Directions L2 Student Resource: Lesson 3 Student Directions L3 Student Graphs: Glycerol content of worms; A, B, C and D L4 (each group needs only one graph per group, not all four) Student Resource: Universal Genetic Code L5 Student Resource: Assessment Instructions Assessment Student Resource: Worm Plates (cannot be reused) Assessment Student Resource: Claim and evidence card Assessment Worm Plates, per lab group One plate of wild type worms per lab group L1, L2 One plate of mutant worms L1, L2 Two plates containing low salt (0.05 M) L2, L3, L4 Two plates containing high salt (0.40 M) L2, L3, L4 Lab Supplies, per lab group Dissecting microscope L1, L2, L3, L4 Disposable gloves, one pair per student L1, L2, L3, L4 Plastic sheet with 4 mm x 4 mm grid L1 ©2013 University of Washington. -

General Lab Instruments 1
General Lab Instruments 1. Pipette HS-TP Series Variable Volume micropipettes HS-TF series precision Fixed Volume Micropipette HS-TS Series Variable Volume micropipettes HS-TS1M Laboratory Straw Micropipette HS-TP Series Multichannel Variable Volume Micropipettes HS-TS Series Multichannel Variable Volume Micropipettes HSG-269 Set of 3 Pipette Pumps: HS-TL series Laboratory Manual Pipette Controller 2ml Blue, 10ml Green, 25ml Red HSG-276 High Precision Professional Scientific Lab Single HSG-275 Single Channel Pipette Multi-Volume Channel Adjust volume Autoclavable Pipette HSG-267 5ml Glass Graduated Pipette Dropper HSG-268 Thick Glass Graduated Dropper Pipettes 10ml HSG-266 1ml Glass Graduated Pipette Dropper HSG-265 Glass Pipette Dropper with Red Rubber Cap HSG-263 3ml Disposable Plastic Eye Dropper HSG-262 1ml Plastic Transfer Pipette HSCG1631 Laboratory Clear Glass Measuring Pipette HS-TEL series Motoirze Pipette Controller 2. Clamps HSG-145 zinc Alloy powder coating German Style Cross Clip Clamp HSG-143 Glass product Laboratory Equipment Cross Clamp HSG-135 Swivel iron plastic coated and brass universal CLAMP HSG-129 Laboratory Double adjustable Swivel clamp HSG-119 Big size Two prongs electroplating extension universal clamp HSG-127 Small Size Three Finger Clamp HSG-153 Lab Brass Cross Clip Clamp Holder HSG-148 UK style Aluminium clamp holder Cross Clamp Holder HSG-163 Multiple Direction Swivel Clamp Holders HSG-139 Laboratory Beaker Chain Clamp HSG-124 LABORATORY TWO PRONG SWIVEL CLAMP HSG-101 Lab Aluminum Flask Clamp HSG-1116 Lab -

CHAPTER 2-1 LABORATORY TECHNIQUES: EQUIPMENT Janice M
Glime, J. M. and Wagner, D. H. 2017. Laboratory Techniques: Equipment. Chapt. 2-1. In: Glime, J. M. Bryophyte Ecology. Volume 2-1-1 3. Methods. Ebook sponsored by Michigan Technological University and the International Association of Bryologists. Ebook last updated 10 April 2021 and available at <http://digitalcommons.mtu.edu/bryophyte-ecology/>. CHAPTER 2-1 LABORATORY TECHNIQUES: EQUIPMENT Janice M. Glime and David H. Wagner TABLE OF CONTENTS Lab Bench Setup ................................................................................................................................................. 2-1-2 Microscopes ........................................................................................................................................................ 2-1-3 Parfocal Adjustment..................................................................................................................................... 2-1-5 Procedure.............................................................................................................................................. 2-1-5 Microscope Use........................................................................................................................................... 2-1-5 Adjusting Light and Learning to Focus................................................................................................ 2-1-5 Adjusting the Focus and Ocular Distance............................................................................................. 2-1-6 Adjustments for Glasses...................................................................................................................... -

Making Biodiesel
Summer Academy 2013 Making Biodiesel Description: There are many benefits of using biofuel as an alternative to petroleum-based fuel. Biodiesel is a renewable fuel made from biologically based oils (vegetable, canola, soybeans etc…) and can be used to power a diesel engine. The chemistry involved in making small scale biodiesel is an easy process that can be done with a few simple chemicals. In today’s lesson, we will be doing background on biodiesel, making a batch of biodiesel and testing it for viscosity, Objectives: x Students will compare physical and chemical properties of vegetable oils x Students will explore the background of biodiesel and look at the economical and environmental benefits of biodiesel x Student will demonstrate the process of making biodiesel on a small scale Standards covered: x 9-10.2.2 Use appropriate safety equipment and precautions during investigations (e.g., goggles, apron, eye wash station) x 9-10.2.7 Maintain clear and accurate records of scientific investigations x 9-10.3.2 Classify changes in matter as physical or chemical Session Organization 9:00-9:30 Cultural connection and general organization 9:30-10:00 Background information 10:00-11:00 Internet activity and discussion finding background on biodiesel 11:00-12:00 Activity making biodiesel 12:00-12:30 Lunch 12:30-1:30 Activity: comparing density and viscosity 1:30-2:30 Activity: calculating heat content in biodiesel vs. diesel 2:30-3:00 Wrap up 1 Activity One: Background information on Biodiesel Using the internet, find the definitions for the -

Chemistry of Combustion Grade/Subject: 8Th Science
Chemistry of Combustion Grade/Subject: 8th Science Strand/Standard 8.1.3 Plan and conduct an investigation and then analyze and interpret the data to identify patterns in changes in a substance’s properties to determine whether a chemical reaction has occurred. Examples could include changes in properties such as color, density, flammability, odor, solubility, or state. (PS1.A, PS1.B) Lesson Performance Expectations: Students will be able to explain the concept of combustion-burning of materials Materials: ● Demonstration/investigation materials: ● 1 Magnesium strip or coil ● 1 Bunsen burner and tubing ● 1 tub with soapy water ● Student investigation materials: ● large jar or beaker ● Combustibles: marshmallows, crackers, peanuts, paper, candle, cardboard etc. ● Lighter or wooden splint (the teacher may wish to do the lighting if you do not want students to have lighters) ● Long tweezers, dissecting probes or another method of holding the burning object ● A tin pie pan for burning items to drop into or be placed into ● A tub of water for emergency flame stoppage Time: 1-2 50 minute periods Teacher Background Information: Chemical reactions with knowledge of reactants and products (bonds are broken and formed) Combustion reactions-Reactions involving the burning of fossil fuels which releases energy for doing work Student Background Knowledge: Students do not need to know about chemical reactions in detail. A short introduction will be sufficient. They need to know what a chemical reaction is and how they work on paper in terms of reactants and products. They will need to be introduced to the concept of combustion-burning fuel to release energy. -

Appendix C a Complete List of Lab Supplies
APPENDIX C A COMPLETE LIST OF LAB SUPPLIES Items in boldface, blue type are found in the laboratory equipment set that is sold with the course. The other materials are available at supermarkets, hardware stores, or drug stores. Module #1 • Safety goggles • A meterstick (A yardstick will work as well; a 12-inch ruler is not long enough.) • Two 8-inch or larger balloons • Two pieces of string long enough to tie the balloons to the meterstick • Tape • A tall glass • A paper towel • A sink full of water • Book (not oversized) • Metric/English ruler or rulers • Water • Vegetable oil • Graduated cylinder or measuring cups • Maple syrup (Natural syrup does not work as well as something like Mrs. Butterworth’s.) • Mass scale Module #2 • Safety goggles • Thermometer • 250 mL beaker (A small glass container will do, but make sure it is safe for boiling water.) • Water (preferably distilled water, which can be purchased at any supermarket) • Ice (preferably crushed) • Alcohol burner or something like a hot plate or stove • Mass scale • Two Styrofoam cups • A chunk of metal that has mass of at least 30 grams (a lead sinker or a very large steel nut, for example) • Boiling water (either in a pot or a beaker) • Kitchen tongs 586 Exploring Creation With Chemistry Module #3 • Safety goggles • 100-mL beaker (A juice glass can be used instead.) • 250-mL beaker (A juice glass can be used instead.) • Watch glass (A small saucer can be used instead. It must cover the mouth of the 100-mL beaker or juice glass listed above.) • Teaspoon • Lye (This is commonly sold in supermarkets with the drain cleaners. -

Everything That You Wanted to Know About Climate Change but Could Not Find in a Text Book!’
‘Everything That You Wanted To Know About Climate Change But Could Not Find In A Text Book!’ ‐O‐O‐O‐ Resources that will assist South African science teachers teaching about the Atmosphere and Climate Change. Produced as part of the SEEDS tour 2011 The Authors Dudley Shallcross is the Professor in Atmospheric Chemistry and Tim Harrison is the School Teacher Fellow at the School of Chemistry at the University of Bristol, UK. The latter is a position for a secondary-school teacher that was created to bridge the gap between secondary schools and universities, and to use the resources of the School of Chemistry to promote chemistry regionally, nationally and internationally. For more information about modelling climate change or about the post of School Teacher Fellow please contact Dudley Shallcross ([email protected]) or Tim Harrison ([email protected]). 1 Contents The Authors ............................................................................................................................ 1 Acknowledgement ................................................................................................................... 2 Practical demonstrations to augment climate change lessons ............................................... 3 Fuelling interest: climate change experiments ........................................................................ 8 Looking to the heavens: climate change experiments .......................................................... 16 A hole in the sky ...................................................................................................................