Unraveling Proteome Changes and Potential Regulatory Proteins Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -
Eif4gi Cooperation in Start Codon Selection
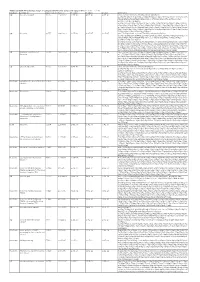
Article Translational Tolerance of Mitochondrial Genes to Metabolic Energy Stress Involves TISU and eIF1- eIF4GI Cooperation in Start Codon Selection Graphical Abstract Authors Hadar Sinvani, Ora Haimov, ..., Benoit Viollet, Rivka Dikstein Correspondence [email protected] In Brief The regulatory element TISU is a translation initiator of short 50 UTR mRNAs. Sinvani et al. report that TISU is enriched in mitochondrial genes and confers resistance to translation inhibition mediated by AMPK and energy deprivation, allowing cells to cope with the stress. TISU-mediated initiation involves cap complex eviction and eIF1- eIF4G1 cooperation. Highlights 0 d TISU, translation initiator of short 5 UTR, is enriched among mitochondrial genes d TISU-mediated initiation involves eIF1-eIF4G1 cooperation and cap complex eviction d Low energy induces a stress response that includes inhibition of global translation d TISU-directed translation enables continuous translation under energy stress Sinvani et al., 2015, Cell Metabolism 21, 479–492 March 3, 2015 ª2015 Elsevier Inc. http://dx.doi.org/10.1016/j.cmet.2015.02.010 Cell Metabolism Article Translational Tolerance of Mitochondrial Genes to Metabolic Energy Stress Involves TISU and eIF1-eIF4GI Cooperation in Start Codon Selection Hadar Sinvani,1,4 Ora Haimov,1,4 Yuri Svitkin,2 Nahum Sonenberg,2 Ana Tamarkin-Ben-Harush,1 Benoit Viollet,3 and Rivka Dikstein1,* 1Department of Biological Chemistry, The Weizmann Institute of Science, Rehovot 76100, Israel 2Department of Biochemistry and Goodman Cancer Centre, McGill University, Montreal, QC H3A 1A3, Canada 3University Paris Descartes, Institut Cochin, 75014 Paris, France 4Co-first author *Correspondence: [email protected] http://dx.doi.org/10.1016/j.cmet.2015.02.010 SUMMARY 2010; Ja¨ ger et al., 2007; Lin et al., 2005; O’Neill et al., 2011; Winder et al., 2000). -

Global Analysis of Trna and Translation Factor Expression Reveals a Dynamic Landscape of Translational Regulation in Human Cancers
ARTICLE https://doi.org/10.1038/s42003-018-0239-8 OPEN Global analysis of tRNA and translation factor expression reveals a dynamic landscape of translational regulation in human cancers Zhao Zhang1, Youqiong Ye1, Jing Gong1, Hang Ruan1, Chun-Jie Liu2, Yu Xiang1, Chunyan Cai3, An-Yuan Guo2, 1234567890():,; Jiqiang Ling4, Lixia Diao5, John N. Weinstein5 & Leng Han 1,6 The protein translational system, including transfer RNAs (tRNAs) and several categories of enzymes, plays a key role in regulating cell proliferation. Translation dysregulation also contributes to cancer development, though relatively little is known about the changes that occur to the translational system in cancer. Here, we present global analyses of tRNAs and three categories of enzymes involved in translational regulation in ~10,000 cancer patients across 31 cancer types from The Cancer Genome Atlas. By analyzing the expression levels of tRNAs at the gene, codon, and amino acid levels, we identified unequal alterations in tRNA expression, likely due to the uneven distribution of tRNAs decoding different codons. We find that overexpression of tRNAs recognizing codons with a low observed-over-expected ratio may overcome the translational bottleneck in tumorigenesis. We further observed overall overexpression and amplification of tRNA modification enzymes, aminoacyl-tRNA synthe- tases, and translation factors, which may play synergistic roles with overexpression of tRNAs to activate the translational systems across multiple cancer types. 1 Department of Biochemistry and Molecular Biology, McGovern Medical School at The University of Texas Health Science Center at Houston, Houston, TX 77030, USA. 2 Department of Bioinformatics and Systems Biology, Hubei Bioinformatics and Molecular Imaging Key Laboratory, Key Laboratory of Molecular Biophysics of the Ministry of Education, College of Life Science and Technology, Huazhong University of Science and Technology Wuhan, 430074 Hubei, People’s Republic of China. -

Identification of Dysregulated Genes in Rheumatoid Arthritis Based on Bioinformatics Analysis
Identification of dysregulated genes in rheumatoid arthritis based on bioinformatics analysis Ruihu Hao1,*, Haiwei Du2,*, Lin Guo1, Fengde Tian1, Ning An1, Tiejun Yang3, Changcheng Wang1, Bo Wang1 and Zihao Zhou1 1 Department of Orthopedics, Affiliated Zhongshan Hospital of Dalian University, Dalian, China 2 Department of Bioinformatics, Beijing Medintell Biomed Co., Ltd, Beijing, China 3 Department of Orthopedics, Affiliated Hospital of BeiHua University, Jilin, China * These authors contributed equally to this work. ABSTRACT Background. Rheumatoid arthritis (RA) is a chronic auto-inflammatory disorder of joints. The present study aimed to identify the key genes in RA for better understanding the underlying mechanisms of RA. Methods. The integrated analysis of expression profiling was conducted to identify differentially expressed genes (DEGs) in RA. Moreover, functional annotation, protein– protein interaction (PPI) network and transcription factor (TF) regulatory network construction were applied for exploring the potential biological roles of DEGs in RA. In addition, the expression level of identified candidate DEGs was preliminarily detected in peripheral blood cells of RA patients in the GSE17755 dataset. Quantitative real-time polymerase chain reaction (qRT-PCR) was conducted to validate the expression levels of identified DEGs in RA. Results. A total of 378 DEGs, including 202 up- and 176 down-regulated genes, were identified in synovial tissues of RA patients compared with healthy controls. DEGs were significantly enriched in -

Variation in Protein Coding Genes Identifies Information Flow
bioRxiv preprint doi: https://doi.org/10.1101/679456; this version posted June 21, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Animal complexity and information flow 1 1 2 3 4 5 Variation in protein coding genes identifies information flow as a contributor to 6 animal complexity 7 8 Jack Dean, Daniela Lopes Cardoso and Colin Sharpe* 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 Institute of Biological and Biomedical Sciences 25 School of Biological Science 26 University of Portsmouth, 27 Portsmouth, UK 28 PO16 7YH 29 30 * Author for correspondence 31 [email protected] 32 33 Orcid numbers: 34 DLC: 0000-0003-2683-1745 35 CS: 0000-0002-5022-0840 36 37 38 39 40 41 42 43 44 45 46 47 48 49 Abstract bioRxiv preprint doi: https://doi.org/10.1101/679456; this version posted June 21, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Animal complexity and information flow 2 1 Across the metazoans there is a trend towards greater organismal complexity. How 2 complexity is generated, however, is uncertain. Since C.elegans and humans have 3 approximately the same number of genes, the explanation will depend on how genes are 4 used, rather than their absolute number. -

(B6;129.Cg-Gt(ROSA)26Sor Tm20(CAG-Ctgf-GFP)Jsd) Were Crossed with Female Foxd1cre/+ Heterozygote Mice 1, and Experimental Mice Were Selected As Foxd1cre/+; Rs26cig/+
Supplemental Information SI Methods Animal studies Heterozygote mice (B6;129.Cg-Gt(ROSA)26Sor tm20(CAG-Ctgf-GFP)Jsd) were crossed with female Foxd1Cre/+ heterozygote mice 1, and experimental mice were selected as Foxd1Cre/+; Rs26CIG/+. In some studies Coll-GFPTg or TCF/Lef:H2B-GFPTg mice or Foxd1Cre/+; Rs26tdTomatoR/+ mice were used as described 2; 3. Left kidneys were subjected to ureteral obstruction using a posterior surgical approach as described 2. In some experiments recombinant mouse DKK1 (0.5mg/kg) or an equal volume of vehicle was administered by daily IP injection. In the in vivo ASO experiment, either specific Lrp6 (TACCTCAATGCGATTT) or scrambled negative control ASO (AACACGTCTATACGC) (30mg/kg) (Exiqon, LNA gapmers) was administered by IP injection on d-1, d1, d4, and d7. In other experiments anti-CTGF domain-IV antibodies (5mg/kg) or control IgG were administered d-1, d1 and d6. All animal experiments were performed under approved IACUC protocols held at the University of Washington and Biogen. Recombinant protein and antibody generation and characterization Human CTGF domain I (sequence Met1 CPDEPAPRCPAGVSLVLDGCGCCRVCAKQLGELCTERDPCDPHKGLFC), domain I+II (sequence Met1CPDEPAPRCPAGVSLVLDGCGCCRVCAKQLGELCTERDPCDPHKGLFCCIFGGT VYRSGESFQSSCKYQCTCLDGAVGCMPLCSMDVRLPSPDCPFPRRVKLPGKCCEE) were cloned and expressed in 293 cells, and purified by Chelating SFF(Ni) Column, tested for single band by SEC and PAGE, and tested for absence of contamination. Domain-IV (sequence GKKCIRTPKISKPIKFELSGCTSMKTYRAKFCGVCTDGRCCTPHRTTTLPVEFKCPDGE VMKKNMMFIKTCACHYNCPGDNDIFESLYYRKMY) was purchased from Peprotech. Mouse or human DKK1 was generated from the coding sequence with some modifications and a tag. Secreted protein was harvested from 293 cells, and purified by nickel column, and tested for activity in a supertopflash (STF) assay 4. DKK1 showed EC50 of 0.69nM for WNT3a-induced WNT signaling in STF cells. -

Bioinformatics Analysis for Multiple Gene Expression Profiles in Sepsis
DATABASE ANALYSIS e-ISSN 1643-3750 © Med Sci Monit, 2020; 26: e920818 DOI: 10.12659/MSM.920818 Received: 2019.10.19 Accepted: 2020.01.21 Bioinformatics Analysis for Multiple Gene Available online: 2020.02.17 Published: 2020.04.13 Expression Profiles in Sepsis Authors’ Contribution: B Jianhua Zhai Department of Emergency, Tianjin Medical University General Hospital, Tianjin, Study Design A C Anlong Qi P.R. China Data Collection B Statistical Analysis C D Yan Zhang Data Interpretation D E Lina Jiao Manuscript Preparation E F Yancun Liu Literature Search F Funds Collection G A Songtao Shou Corresponding Author: Songtao Shou, e-mail: [email protected] Source of support: This work was supported by the National Natural Science Foundation of China (Grant No. 81871593) Background: This work aimed to screen key biomarkers related to sepsis progression by bioinformatics analyses. Material/Methods: The microarray datasets of blood and neutrophils from patients with sepsis or septic shock were download- ed from Gene Expression Omnibus database. Then, differentially expressed genes (DEGs) from 4 groups (sep- sis versus normal blood samples; septic shock versus normal blood samples; sepsis neutrophils versus normal controls and septic shock neutrophils versus controls) were respectively identified followed by functional anal- yses. Subsequently, protein–protein network was constructed, and key functional sub-modules were extracted. Finally, receiver operating characteristic analysis was conducted to evaluate diagnostic values of key genes. Results: There were 2082 DEGs between blood samples of sepsis patients and controls, 2079 DEGs between blood sam- ples of septic shock patients and healthy individuals, 6590 DEGs between neutrophils from sepsis and con- trols, and 1056 DEGs between neutrophils from septic shock patients and normal controls. -
Additional Tables.Xlsx
Additional Table 5 Enriched pathways of upregulated DEGs after spinal cord injury in WT (P < 0.05, q < 0.05) Database Description Ratio of DEGs Ratio of P -value P adjust q value gene name GO ribosomal subunit 87/755 214/12262 1.03E-49 4.61E-47 4.20E-47 Gm6576/Gm10036/Gm5786/Mrps28/Gm9493/Rpl13- ps3/Rpl7l1/Mrpl10/Mrpl44/Rps20/Mrpl51/Mrps11/Mpv17l2/Mrps7/Gm10020/Gm10073 /Mrpl2/Mrpl52/Rpl22/Mrpl4/Mrps24/Zcchc17/Rpl28/Mrpl34/Rpl29/Mrps36/Rpl9- ps6/Hba-a2/Hba-a1/Rpl23a- ps3/Rpl15/Rack1/Mrps18a/Mrpl28/Rpl10a/Mrpl12/Rpl18/Rpl35a/Mrpl41/Mrps12/Rpl36 a/Rps9/Rps18/Rps5/Rps27a/Rps24/Rplp2/Rpl14/Mrpl30/Rps6/Rpl6/Rps3/Rps4x/Rpsa/R pl7/Rpl27a/Rps23/Rps10/Mrps21/Rpl36/Rpl30/Rps17/Mrpl42/Rpl35/Rpl27/Rplp0/Rps28 /Rpl31/Rpl39/Rps13/Rpl13/Rps11/Rpl8/Rps3a1/Fau/Rpl9/Rpl3/Rpl26/Rps15/Rpl13a/Uba 52/Rpl24/Rpl19/Rps14/Rpl4/Rps27/Rplp1 GO ribosome 93/755 248/12262 1.67E-49 4.61E-47 4.20E-47 Gm6576/Pnpt1/Gm10036/Gm5786/Mrps28/Gm9493/Rpl13- ps3/Rpl7l1/Mrpl10/Mrpl44/Rps20/Mrpl51/Mrps11/Mpv17l2/Mrps7/Gm10020/Gm10073 /Mrpl2/Mrpl52/Rpl22/Mrpl4/Mrps24/Zcchc17/Rpl28/Mrpl34/Rpl29/Mrps36/Rpl9- ps6/Hba-a2/Hba-a1/Rpl23a- ps3/Rpl15/Rack1/Eif3h/Mrps18a/Mrpl28/Mrps33/Rpl10a/Mrpl12/Rpl18/Rpl35a/Btf3/Mrp l41/Mrps12/Rpl36a/Rps9/Rps18/Rps5/Rps27a/Rps24/Rplp2/Rpl14/Mrpl30/Rps6/Rpl6/R ps3/Rps4x/Rpsa/Rpl7/Rpl27a/Rps23/Rps10/Mrps21/Rpl36/Rpl30/Rps17/Mrpl42/Rpl35/R pl27/Rplp0/Rps28/Rpl31/Rpl39/Rps13/Rpl13/Rps11/Rpl8/Rps3a1/Fau/Rpl9/Rpl3/Rpl26/ Rps15/Rpl13a/Uba52/Rpl41/Rpl24/Rpl19/Rps14/Rpl4/Ndufa7/Rps27/Rplp1 GO structural constituent of 76/733 169/11958 1.65E-47 1.16E-44 1.12E-44 Gm6576/Gm10036/Gm5786/Rpl7l1/Mrpl10/Rps20/Mrpl51/Mrps11/Mrps7/Gm10020/G -

Glucagon-Like Peptide-1 Receptor Agonists Increase Pancreatic Mass by Induction of Protein Synthesis
Page 1 of 73 Diabetes Glucagon-like peptide-1 receptor agonists increase pancreatic mass by induction of protein synthesis Jacqueline A. Koehler1, Laurie L. Baggio1, Xiemin Cao1, Tahmid Abdulla1, Jonathan E. Campbell, Thomas Secher2, Jacob Jelsing2, Brett Larsen1, Daniel J. Drucker1 From the1 Department of Medicine, Tanenbaum-Lunenfeld Research Institute, Mt. Sinai Hospital and 2Gubra, Hørsholm, Denmark Running title: GLP-1 increases pancreatic protein synthesis Key Words: glucagon-like peptide 1, glucagon-like peptide-1 receptor, incretin, exocrine pancreas Word Count 4,000 Figures 4 Tables 1 Address correspondence to: Daniel J. Drucker M.D. Lunenfeld-Tanenbaum Research Institute Mt. Sinai Hospital 600 University Ave TCP5-1004 Toronto Ontario Canada M5G 1X5 416-361-2661 V 416-361-2669 F [email protected] 1 Diabetes Publish Ahead of Print, published online October 2, 2014 Diabetes Page 2 of 73 Abstract Glucagon-like peptide-1 (GLP-1) controls glucose homeostasis by regulating secretion of insulin and glucagon through a single GLP-1 receptor (GLP-1R). GLP-1R agonists also increase pancreatic weight in some preclinical studies through poorly understood mechanisms. Here we demonstrate that the increase in pancreatic weight following activation of GLP-1R signaling in mice reflects an increase in acinar cell mass, without changes in ductal compartments or β-cell mass. GLP-1R agonists did not increase pancreatic DNA content or the number of Ki67+ cells in the exocrine compartment, however pancreatic protein content was increased in mice treated with exendin-4 or liraglutide. The increased pancreatic mass and protein content was independent of cholecystokinin receptors, associated with a rapid increase in S6 kinase phosphorylation, and mediated through the GLP-1 receptor. -

Transcriptome Profiles of Porcine Oocytes and Their Corresponding Cumulus Cells Reveal Functional Gene Regulatory Networks
Transcriptome profiles of porcine oocytes and their corresponding cumulus cells reveal functional gene regulatory networks by Katelyn Marie Kimble A thesis submitted to the Graduate Faculty of Auburn University in partial fulfillment of the requirements for the Degree of Master of Science Auburn, Alabama August 4, 2018 Keywords: oocyte, cumulus cells, signaling, transcriptome, porcine Approved by Fernando Biase, Chair, Assistant Professor, Dept. of Animal Sciences Paul Dyce, Assistant Professor, Dept. of Animal Sciences Julie Gard, Professor, Dept. of Clinical Sciences Jacek Wower, Professor, Dept. of Animal Sciences Abstract The oocyte acquires developmental competence as it progresses through folliculogenesis. It does so, by communicating with the surrounding cumulus cells in a bidirectional fashion. Analyses of single oocytes are essential for further clarification of these molecular mechanisms. Standard protocols to obtain RNA for single-cell RNA-Seq involve RNA extraction kits that select for mRNAs or cell lysis procedures, which present limitations when the oocyte is the cell of interest. We adapted the phenol-chloroform procedure for the purification of total RNA from single oocytes with modifications that included the use of Phasemaker™ tubes, a second chloroform wash, and the precipitation of the RNA with glycogen in microcentrifuge tubes. We profiled the total RNA of single oocytes (bovine and porcine), and observed distinct peaks for small RNAs, 18S, and 28S. We amplified the total mRNA and observed DNA fragments longer than 5000 bp in length, suitable for single-cell RNA sequencing. We applied this approach to sequence the transcriptome of 17 prepubertal porcine oocytes and their corresponding cumulus cells. We hypothesized that the transcript profiles of the cumulus cells and oocyte display distinct gene regulatory networks within the oocyte and cumulus cells. -

Supplementary Data
SUPPLEMENTAL INFORMATION A study restricted to chemokine receptors as well as a genome-wide transcript analysis uncovered CXCR4 as preferentially expressed in Ewing's sarcoma (Ewing's sarcoma) cells of metastatic origin (Figure 4). Transcriptome analyses showed that in addition to CXCR4, genes known to support cell motility and invasion topped the list of genes preferentially expressed in metastasis-derived cells (Figure 4D). These included kynurenine 3-monooxygenase (KMO), galectin-1 (LGALS1), gastrin-releasing peptide (GRP), procollagen C-endopeptidase enhancer (PCOLCE), and ephrin receptor B (EPHB3). KMO, a key enzyme of tryptophan catabolism, has not been linked to metastasis. Tryptophan and its catabolites, however, are involved in immune evasion by tumors, a process that can assist in tumor progression and metastasis (1). LGALS1, GRP, PCOLCE and EPHB3 have been linked to tumor progression and metastasis of several cancers (2-4). Top genes preferentially expressed in L-EDCL included genes that suppress cell motility and/or potentiate cell adhesion such as plakophilin 1 (PKP1), neuropeptide Y (NPY), or the metastasis suppressor TXNIP (5-7) (Figure 4D). Overall, L-EDCL were enriched in gene sets geared at optimizing nutrient transport and usage (Figure 4D; Supplementary Table 3), a state that may support the early stages of tumor growth. Once tumor growth outpaces nutrient and oxygen supplies, gene expression programs are usually switched to hypoxic response and neoangiogenesis, which ultimately lead to tumor egress and metastasis. Accordingly, gene sets involved in extracellular matrix remodeling, MAPK signaling, and response to hypoxia were up-regulated in M-EDCL (Figure 4D; Supplementary Table 4), consistent with their association to metastasis in other cancers (8, 9). -

SUPPLEMENTARY MATERIALS and METHODS PBMC Transcriptomics
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) Gut SUPPLEMENTARY MATERIALS AND METHODS PBMC transcriptomics identifies immune-metabolism disorder during the development of HBV-ACLF Contents l Supplementary methods l Supplementary Figure 1 l Supplementary Figure 2 l Supplementary Figure 3 l Supplementary Figure 4 l Supplementary Figure 5 l Supplementary Table 1 l Supplementary Table 2 l Supplementary Table 3 l Supplementary Table 4 l Supplementary Tables 5-14 l Supplementary Table 15 l Supplementary Table 16 l Supplementary Table 17 Li J, et al. Gut 2021;0:1–13. doi: 10.1136/gutjnl-2020-323395 BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) Gut SUPPLEMENTARY METHODS Test for HBV DNA The levels of HBV DNA were detected using real-time PCR with a COBAS® AmpliPrep/COBAS® TaqMan 48 System (Roche, Basel, Switzerland) and HBV Test v2.0. Criteria for diagnosing cirrhosis Pathology The gold standard for the diagnosis of cirrhosis is a liver biopsy obtained through a percutaneous or transjugular approach.1 Ultrasonography was performed 2-4 hours before biopsy. Liver biopsy specimens were obtained by experienced physicians. Percutaneous transthoracic puncture of the liver was performed according to the standard criteria. After biopsy, patients were monitored in the hospital with periodic analyses of haematocrit and other vital signs for 24 hours. Cirrhosis was diagnosed according to the globally agreed upon criteria.2 Cirrhosis is defined based on its pathological features under a microscope: (a) the presence of parenchymal nodules, (b) differences in liver cell size and appearance, (c) fragmentation of the biopsy specimen, (d) fibrous septa, and (d) an altered architecture and vascular relationships.