The Human Flavoproteome
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Ferric Reductase Activity of the Arsh Protein from Acidithiobacillus Ferrooxidans
J. Microbiol. Biotechnol. (2011), 21(5), 464–469 doi: 10.4014/jmb.1101.01020 First published online 13 April 2011 Ferric Reductase Activity of the ArsH Protein from Acidithiobacillus ferrooxidans Mo, Hongyu1,2, Qian Chen1,2, Juan Du1, Lin Tang1, Fang Qin1, Bo Miao2, Xueling Wu2, and Jia Zeng1,2* 1College of Biology, Hunan University, Changsha, Hunan 410082, P. R. China 2Department of Bioengineering, Central South University, Changsha, Hunan 410083, P. R. China Received: January 14, 2011 / Revised: March 10, 2011 / Accepted: March 11, 2011 The arsH gene is one of the arsenic resistance system in iron (free or chelated) into ferrous iron before its incorporation bacteria and eukaryotes. The ArsH protein was annotated into heme and nonheme iron-containing proteins. Ferric as a NADPH-dependent flavin mononucleotide (FMN) reductase catalyzes the reduction of complexed Fe3+ to reductase with unknown biological function. Here we complexed Fe2+ using NAD(P)H as the electron donor. The report for the first time that the ArsH protein showed resulting Fe2+ is subsequently released and incorporated high ferric reductase activity. Glu104 was an essential into iron-containing proteins [17]. residue for maintaining the stability of the FMN cofactor. Here we report for the first time that the ArsH protein The ArsH protein may perform an important role for showed high ferric reduction activity. The ArsH from A. cytosolic ferric iron assimilation in vivo. ferrooxidans may perform an important role as a NADPH- Keywords: Acidithiobacillus ferrooxidans, ArsH, flavoprotein, dependent ferric reductase for cytosolic ferric iron assimilation ferric reductase in vivo. MATERIALS AND METHODS Arsenic resistance genes are widespread in nature. -

Progressive Encephalopathy and Central Hypoventilation Related to Homozygosity of NDUFV1 Nuclear Gene, a Rare Mitochondrial Disease
Avens Publishing Group Inviting Innovations Open Access Case Report J Pediatr Child Care August 2019 Volume:5, Issue:1 © All rights are reserved by AL-Buali MJ, et al. AvensJournal Publishing of Group Inviting Innovations Progressive Encephalopathy Pediatrics & and Central Hypoventilation Child Care AL-Buali MJ*, Al Ramadhan S, Al Buali H, Al-Faraj J and Related to Homozygosity of Al Mohanna M Pediatric Department , Maternity Children Hospital , Saudi Arabia *Address for Correspondence: NDUFV1 Nuclear Gene, a Rare Al-buali MJ, Pediatric Consultant and Consultant of Medical Genetics, Deputy Chairman of Medical Genetic Unite, Pediatrics Department , Maternity Children Hospital, Al-hassa, Hofuf city, Mitochondrial Disease Saudi Arabia; E-mail: [email protected] Submission: 15 July 2019 Accepted: 5 August 2019 Keywords: Progressive encephalopathy; Central hypoventilation; Published: 9 August 2019 Nuclear mitochondrial disease; NDUFV1 gene Copyright: © 2019 AL-Buali MJ, et al. This is an open access article distributed under the Creative Commons Attribution License, which Abstract permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Background: Mitochondrial diseases are a group of disorders caused by dysfunctional organelles that generate energy for our body. Mitochondria small double-membrane organelles found in of the most common groups of genetic diseases with a minimum every cell of the human body except red blood cells. Mitochondrial diseases are sometimes caused by mutations in the mitochondrial DNA prevalence of greater than 1 in 5000 in adults. Mitochondrial diseases that affect mitochondrial function. Other mitochondrial diseases are can be present at birth but can be manifested also at any age [2]. -

In Vitro Treatment of Hepg2 Cells with Saturated Fatty Acids Reproduces
© 2015. Published by The Company of Biologists Ltd | Disease Models & Mechanisms (2015) 8, 183-191 doi:10.1242/dmm.018234 RESEARCH ARTICLE In vitro treatment of HepG2 cells with saturated fatty acids reproduces mitochondrial dysfunction found in nonalcoholic steatohepatitis Inmaculada García-Ruiz1,*, Pablo Solís-Muñoz2, Daniel Fernández-Moreira3, Teresa Muñoz-Yagüe1 and José A. Solís-Herruzo1 ABSTRACT INTRODUCTION Activity of the oxidative phosphorylation system (OXPHOS) is Nonalcoholic fatty liver disease (NAFLD) represents a spectrum of decreased in humans and mice with nonalcoholic steatohepatitis. liver diseases extending from pure fatty liver through nonalcoholic Nitro-oxidative stress seems to be involved in its pathogenesis. The steatohepatitis (NASH) to cirrhosis and hepatocarcinoma that occurs aim of this study was to determine whether fatty acids are implicated in individuals who do not consume a significant amount of alcohol in the pathogenesis of this mitochondrial defect. In HepG2 cells, we (Matteoni et al., 1999). Although the pathogenesis of NAFLD analyzed the effect of saturated (palmitic and stearic acids) and remains undefined, the so-called ‘two hits’ model of pathogenesis monounsaturated (oleic acid) fatty acids on: OXPHOS activity; levels has been proposed (Day and James, 1998). Whereas the ‘first hit’ of protein expression of OXPHOS complexes and their subunits; gene involves the accumulation of fat in the liver, the ‘second hit’ expression and half-life of OXPHOS complexes; nitro-oxidative stress; includes oxidative stress resulting in inflammation, stellate cell and NADPH oxidase gene expression and activity. We also studied the activation, fibrogenesis and progression of NAFLD to NASH effects of inhibiting or silencing NADPH oxidase on the palmitic-acid- (Chitturi and Farrell, 2001). -
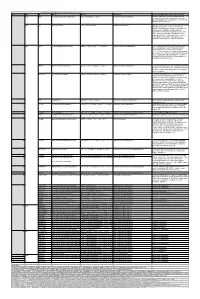
When the Reaction Is
Table S3. iJL1678-ME model modification (blocked reactions) Iter. Cat. ID Name Formula Subsystem Comments (When the reaction is turned on) 1 bp2 EDD 6-phosphogluconate dehydratase 6pgc_c⇌2ddg6p_c + h2o_c Pentose Phosphate Pathway Create a major effect of steep acetate overflow elevation in high growth. Comparing to the main glycolytic pathway, it is metabolicly less efficient but proteomicly more efficient. bp1 ICL Isocitrate lyase icit_c→glx_c + succ_c Anaplerotic Reactions Bypass for the main TCA cycle pathways from turning isocitrate to succinate, when ICL is turned on, Isocitrate dehydrogenase(ICDHyr), 2-Oxogluterate dehydrogenase(AKGDH) and Succinyl-CoA synthetase (ATP-forming,SUCOAS) would reduce. Ref. (1) and (2) shows that this reaction is off in higher growth. Ref. (3) shows that this reaction is converging to being off when the dynamic of respiration using enzyme kinetics is simulated. 2 bp1 ABTA 4-aminobutyrate transaminase 4abut_c + akg_c⇌glu__L_c + sucsal_c Arginine and Proline Metabolism Another backup pathway of succinate production, from 2-Oxoglutarate (akg). Respiration would be induced when it is on, since the flux through ETC(CYTBO3_4pp and ATPS4rpp) would increase. As it requires the co-factor pyridoxal 5'-phosphate(2−) to get catalyzed(4), indicating that this reaction is regulated by the flux of other reactions(pyridoxal 5'- phosphate(2-) production, etc.). 3 GLYAT Glycine C-acetyltransferase accoa_c + gly_c⇌2aobut_c + coa_c Glycine and Serine Metabolism A reaction that back up for the respiration. Reactions fluxes in TCA cycle would drop when this reaction is turned on. It also requires pyridoxal 5'-phosphate(2−) for the regulation. 4 NADTRHD NAD transhydrogenase nad_c + nadph_c⇌nadh_c + nadp_c Oxidative Phosphorylation A reaction that would make the transition between NAD and NADP metabolically more efficient. -
Generate Metabolic Map Poster
Authors: Pallavi Subhraveti Anamika Kothari Quang Ong Ron Caspi An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Peter D Karp Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Csac1394711Cyc: Candidatus Saccharibacteria bacterium RAAC3_TM7_1 Cellular Overview Connections between pathways are omitted for legibility. Tim Holland TM7C00001G0420 TM7C00001G0109 TM7C00001G0953 TM7C00001G0666 TM7C00001G0203 TM7C00001G0886 TM7C00001G0113 TM7C00001G0247 TM7C00001G0735 TM7C00001G0001 TM7C00001G0509 TM7C00001G0264 TM7C00001G0176 TM7C00001G0342 TM7C00001G0055 TM7C00001G0120 TM7C00001G0642 TM7C00001G0837 TM7C00001G0101 TM7C00001G0559 TM7C00001G0810 TM7C00001G0656 TM7C00001G0180 TM7C00001G0742 TM7C00001G0128 TM7C00001G0831 TM7C00001G0517 TM7C00001G0238 TM7C00001G0079 TM7C00001G0111 TM7C00001G0961 TM7C00001G0743 TM7C00001G0893 TM7C00001G0630 TM7C00001G0360 TM7C00001G0616 TM7C00001G0162 TM7C00001G0006 TM7C00001G0365 TM7C00001G0596 TM7C00001G0141 TM7C00001G0689 TM7C00001G0273 TM7C00001G0126 TM7C00001G0717 TM7C00001G0110 TM7C00001G0278 TM7C00001G0734 TM7C00001G0444 TM7C00001G0019 TM7C00001G0381 TM7C00001G0874 TM7C00001G0318 TM7C00001G0451 TM7C00001G0306 TM7C00001G0928 TM7C00001G0622 TM7C00001G0150 TM7C00001G0439 TM7C00001G0233 TM7C00001G0462 TM7C00001G0421 TM7C00001G0220 TM7C00001G0276 TM7C00001G0054 TM7C00001G0419 TM7C00001G0252 TM7C00001G0592 TM7C00001G0628 TM7C00001G0200 TM7C00001G0709 TM7C00001G0025 TM7C00001G0846 TM7C00001G0163 TM7C00001G0142 TM7C00001G0895 TM7C00001G0930 Detoxification Carbohydrate Biosynthesis DNA combined with a 2'- di-trans,octa-cis a 2'- Amino Acid Degradation an L-methionyl- TM7C00001G0190 superpathway of pyrimidine deoxyribonucleotides de novo biosynthesis (E. -

Reaction Mechanism of Azoreductases Suggests Convergent Evolution with Quinone Oxidoreductases
Protein Cell 2010, 1(8): 780–790 Protein & Cell DOI 10.1007/s13238-010-0090-2 RESEARCH ARTICLE Reaction mechanism of azoreductases suggests convergent evolution with quinone oxidoreductases ✉ Ali Ryan*, Chan-Ju Wang*, Nicola Laurieri*, Isaac Westwood, Edith Sim Department of Pharmacology, University of Oxford, Mansfield Road, Oxford, OX1 3QT, UK ✉ Correspondence: [email protected] Received June 2, 2010 Accepted June 28, 2010 ABSTRACT the dye used for dying fabrics does not bind to the fabric, and as a result, is lost in the effluent (Shore, 1995). As these dyes Azoreductases are involved in the bioremediation by can be carcinogenic (Alves de Lima et al., 2007) their removal bacteria of azo dyes found in waste water. In the gut flora, from the effluent is essential. This has driven investigations they activate azo pro-drugs, which are used for treatment into both electrochemical reduction of azo dyes (Wang et al., of inflammatory bowel disease, releasing the active 2010b), as well as the use of both aerobic and anaerobic component 5-aminosalycilic acid. The bacterium bacteria for bioremediation (dos Santos et al., 2007; You et P. aeruginosa has three azoreductase genes, paAzoR1, al., 2007). paAzoR2 and paAzoR3, which as recombinant enzymes Azo bonds are also found in some drugs including those have been shown to have different substrate specifici- used for the treatment of inflammatory bowel disease (IBD). ties. The mechanism of azoreduction relies upon tauto- Azo compounds have also been shown to be effective merisation of the substrate to the hydrazone form. We antibacterial (Farghaly and Abdalla, 2009) and antitumor (El- report here the characterization of the P. -

(12) Patent Application Publication (10) Pub. No.: US 2017/0137846A1 ATSUMI Et Al
US 2017.0137846A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2017/0137846A1 ATSUMI et al. (43) Pub. Date: May 18, 2017 (54) BACTERIA ENGINEERED FOR Related U.S. Application Data CONVERSION OF ETHYLENE TO (60) Provisional application No. 62/009,857, filed on Jun. N-BUTANOL 9, 2014. (71) Applicant: The Regents of the University of California, Oakland, CA (US) Publication Classification (51) Int. Cl. (72) Inventors: Shota ATSUMI, Davis, CA (US); CI2P 7/06 (2006.01) Michael D. TONEY, Davis, CA (US); CI2P 7/16 (2006.01) Gabriel M. RODRIGUEZ, Davis, CA CI2N 15/52 (2006.01) (US); Yohei TASHIRO, Davis, CA (52) U.S. Cl. (US); Justin B. SIEGEL, Davis, CA CPC ................ CI2P 7/06 (2013.01): CI2N 15/52 (US); D. Alexander CARLIN, Davis, (2013.01); C12Y402/01053 (2013.01); C12Y CA (US); Irina KORYAKINA, Davis, 402/01095 (2013.01); C12Y 114/13 (2013.01); CA (US); Shuchi H. DESAI, Davis, CI2Y 102/01002 (2013.01); C12Y 114/13069 CA (US) (2013.01); C12Y 203/01009 (2013.01); C12Y 101/01157 (2013.01); C12Y402/01 (2013.01); (73) Assignee: The Regents of the University of CI2Y 103/01038 (2013.01); C12Y 102/01003 California, Oakland, CA (US) (2013.01); C12P 7/16 (2013.01) (21) Appl. No.: 15/317,656 (57) ABSTRACT (22) PCT Fed: Jun. 9, 2015 The present disclosure provides recombinant bacteria with elevated production of ethanol and/or n-butanol from eth (86) PCT No.: PCT/US 15/34942 ylene. Methods for the production of the recombinant bac S 371 (c)(1), teria, as well as for use thereof for production of ethanol (2) Date: Dec. -

Q 297 Suppl USE
The following supplement accompanies the article Atlantic salmon raised with diets low in long-chain polyunsaturated n-3 fatty acids in freshwater have a Mycoplasma dominated gut microbiota at sea Yang Jin, Inga Leena Angell, Simen Rød Sandve, Lars Gustav Snipen, Yngvar Olsen, Knut Rudi* *Corresponding author: [email protected] Aquaculture Environment Interactions 11: 31–39 (2019) Table S1. Composition of high- and low LC-PUFA diets. Stage Fresh water Sea water Feed type High LC-PUFA Low LC-PUFA Fish oil Initial fish weight (g) 0.2 0.4 1 5 15 30 50 0.2 0.4 1 5 15 30 50 80 200 Feed size (mm) 0.6 0.9 1.3 1.7 2.2 2.8 3.5 0.6 0.9 1.3 1.7 2.2 2.8 3.5 3.5 4.9 North Atlantic fishmeal (%) 41 40 40 40 40 30 30 41 40 40 40 40 30 30 35 25 Plant meals (%) 46 45 45 42 40 49 48 46 45 45 42 40 49 48 39 46 Additives (%) 3.3 3.2 3.2 3.5 3.3 3.4 3.9 3.3 3.2 3.2 3.5 3.3 3.4 3.9 2.6 3.3 North Atlantic fish oil (%) 9.9 12 12 15 16 17 18 0 0 0 0 0 1.2 1.2 23 26 Linseed oil (%) 0 0 0 0 0 0 0 6.8 8.1 8.1 9.7 11 10 11 0 0 Palm oil (%) 0 0 0 0 0 0 0 3.2 3.8 3.8 5.4 5.9 5.8 5.9 0 0 Protein (%) 56 55 55 51 49 47 47 56 55 55 51 49 47 47 44 41 Fat (%) 16 18 18 21 22 22 22 16 18 18 21 22 22 22 28 31 EPA+DHA (% diet) 2.2 2.4 2.4 2.9 3.1 3.1 3.1 0.7 0.7 0.7 0.7 0.7 0.7 0.7 4 4.2 Table S2. -

Ferroptosis-Related Flavoproteins: Their Function and Stability
International Journal of Molecular Sciences Review Ferroptosis-Related Flavoproteins: Their Function and Stability R. Martin Vabulas Charité-Universitätsmedizin, Institute of Biochemistry, Charitéplatz 1, 10117 Berlin, Germany; [email protected]; Tel.: +49-30-4505-28176 Abstract: Ferroptosis has been described recently as an iron-dependent cell death driven by peroxida- tion of membrane lipids. It is involved in the pathogenesis of a number of diverse diseases. From the other side, the induction of ferroptosis can be used to kill tumor cells as a novel therapeutic approach. Because of the broad clinical relevance, a comprehensive understanding of the ferroptosis-controlling protein network is necessary. Noteworthy, several proteins from this network are flavoenzymes. This review is an attempt to present the ferroptosis-related flavoproteins in light of their involvement in anti-ferroptotic and pro-ferroptotic roles. When available, the data on the structural stability of mutants and cofactor-free apoenzymes are discussed. The stability of the flavoproteins could be an important component of the cellular death processes. Keywords: flavoproteins; riboflavin; ferroptosis; lipid peroxidation; protein quality control 1. Introduction Human flavoproteome encompasses slightly more than one hundred enzymes that par- ticipate in a number of key metabolic pathways. The chemical versatility of flavoproteins relies on the associated cofactors, flavin mononucleotide (FMN) and flavin adenine dinu- cleotide (FAD). In humans, flavin cofactors are biosynthesized from a precursor riboflavin that has to be supplied with food. To underline its nutritional essentiality, riboflavin is called vitamin B2. In compliance with manifold cellular demands, flavoproteins have been accommo- Citation: Vabulas, R.M. dated to operate at different subcellular locations [1]. -

Probing the Flavin Transfer Mechanism in Alkanesulfonate Monooxygenase System
bioRxiv preprint doi: https://doi.org/10.1101/433839; this version posted October 2, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Probing the flavin transfer mechanism in alkanesulfonate monooxygenase system Dayal PV and Ellis HR Department of Chemistry and Biochemistry, Auburn University, Alabama, 36849 Email: [email protected] Abstract Bacteria acquire sulfur through the sulfur assimilation pathway, but under sulfur limiting conditions bacteria must acquire sulfur from alternative sources. The alkanesulfonate monooxygenase enzymes are expressed under sulfur-limiting conditions, and catalyze the desulfonation of wide-range of alkanesulfonate substrates. The SsuE enzyme is an NADPH-dependent FMN reductase that provides reduced flavin to the SsuD monooxygenase. The mechanism for the transfer of reduced flavin in flavin dependent two-component systems occurs either by free- diffusion or channeling. Previous studies have shown the presence of protein-protein interactions between SsuE and SsuD, but the identification of putative interaction sights have not been investigated. Current studies utilized HDX-MS to identify protective sites on SsuE and SsuD. A conserved α-helix on SsuD showed a decrease in percent deuteration when SsuE was included in the reaction. This suggests the role of α-helix in promoting protein-protein interactions. Specific SsuD variants were generated in order to investigate the role of these residues in protein-protein interactions and catalysis. Variant containing substitutions at the charged residues showed a six-fold decrease in the activity, while a deletion variant of SsuD lacking the α-helix showed no activity when compared to wild-type SsuD. -

Beneficial Effects of Resveratrol on Respiratory Chain Defects in Patients' Fibroblas
Beneficial effects of Resveratrol on respiratory chain defects in patients’ fibroblasts involve estrogen receptor and estrogen-related receptor α signaling. Alexandra Lopes Costa1, Carole Le Bachelier1, Lise Mathieu1, Agnès Rotig2, Avihu Boneh3, Pascale De Lonlay2, 4, Mark A. Tarnopolsky5, David R. Thorburn3, Jean Bastin1, 6 and Fatima Djouadi1, 6 * 1 INSERM U747, Université Paris Descartes, Paris, France 2 Université Paris Descartes-Sorbonne Paris Cité, Institut Imagine and INSERM U781, Paris, France 3 Department of Paediatrics, University of Melbourne, Murdoch Children's Research Institute and Royal Children's Hospital, Melbourne, Victoria, Australia 4 Centre de Référence des Maladies Métaboliques, Hôpital Necker, Paris, France 5 Neuromuscular and Neurometabolic Unit, McMaster University, Hamilton, Ontario, Canada 6 Assistance Publique Hôpitaux de Paris, Paris, France * Corresponding author: Fatima Djouadi INSERM U747 Université Paris Descartes UFR Biomédicale des Saints-Pères 45, rue des Saints-Pères 75270 Paris cedex 06 France Phone: 33-1-42862219 Fax: 33-1-42863868 e-mail: [email protected] ABSTRACT Mitochondrial respiratory chain disorders are the most prevalent inborn metabolic diseases and remain without effective treatment to date. Up-regulation of residual enzyme activity has been proposed as a possible therapeutic approach in this group of disorders. Since resveratrol (RSV), a natural compound, was proposed to stimulate mitochondrial metabolism in rodents, we tested the effect of this compound on mitochondrial functions in control or in Complex I (CI)- or Complex IV (CIV)-deficient patients’ fibroblasts. We show that RSV stimulates the expression of a panel of proteins representing structural subunits or assembly factors of the five RC complexes, in control fibroblasts. -

Structural Features That Promote Catalysis in Two-Component Systems Involved in Sulfur Metabolism
Structural Features That Promote Catalysis in Two-Component Systems Involved in Sulfur Metabolism by Richard Allen Hagen A dissertation to the Graduate Faculty of Auburn University in partial fulfillment of the requirements for the Degree of Doctor of Philosophy Auburn, Alabama August 8, 2020 Copyright 2020 by Richard Allen Hagen Approved by Holly R. Ellis, Chair, Professor of Chemistry and Biochemistry Douglas C. Goodwin, Professor of Chemistry and Biochemistry Evert C. Duin, Professor of Chemistry and Biochemistry Steven Mansoorabadi, Associate Professor of Chemistry and Biochemistry Abstract Sulfur is an essential element important in the synthesis of biomolecules. Bacteria are able to assimilate inorganic sulfur for the biosynthesis of L-cysteine. Inorganic sulfate is often unavailable, so bacteria have evolved multiple metabolic pathways to obtain sulfur from alternative sources. Interestingly, many of the enzymes involved in sulfur acquisition are flavin- dependent two-component systems. These two-component systems consist of a flavin reductase and monooxygenase that utilize flavin to cleave the carbon-sulfur bonds of organosulfur compounds. The two-component systems differ in their characterized sulfur substrate specificity. Enzymes SsuE/SsuD are involved in the desulfonation of linear alkanesulfonates (C2-C10), enzymes MsuE/MsuD utilize methanesulfonate (C1), and enzymes SfnF/SfnG utilize DMSO2 as a sulfur source. The flavin reductases involved in sulfur assimilation utilize FMN as a substrate but differ in their ability to utilize NADH or NADPH. The alkanesulfonate monooxygenase system was the first two-component flavin-dependent system expressed during sulfur limiting conditions that was characterized. The flavin reductase (SsuE) and monooxygenase (SsuD) have distinct structural and functional properties, but the two enzymes must synchronize their functions for catalysis to occur.