Categoryterm Count % Pvalue Genes List Total Pop Hitspop
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Analysis of Gene Expression Data for Gene Ontology
ANALYSIS OF GENE EXPRESSION DATA FOR GENE ONTOLOGY BASED PROTEIN FUNCTION PREDICTION A Thesis Presented to The Graduate Faculty of The University of Akron In Partial Fulfillment of the Requirements for the Degree Master of Science Robert Daniel Macholan May 2011 ANALYSIS OF GENE EXPRESSION DATA FOR GENE ONTOLOGY BASED PROTEIN FUNCTION PREDICTION Robert Daniel Macholan Thesis Approved: Accepted: _______________________________ _______________________________ Advisor Department Chair Dr. Zhong-Hui Duan Dr. Chien-Chung Chan _______________________________ _______________________________ Committee Member Dean of the College Dr. Chien-Chung Chan Dr. Chand K. Midha _______________________________ _______________________________ Committee Member Dean of the Graduate School Dr. Yingcai Xiao Dr. George R. Newkome _______________________________ Date ii ABSTRACT A tremendous increase in genomic data has encouraged biologists to turn to bioinformatics in order to assist in its interpretation and processing. One of the present challenges that need to be overcome in order to understand this data more completely is the development of a reliable method to accurately predict the function of a protein from its genomic information. This study focuses on developing an effective algorithm for protein function prediction. The algorithm is based on proteins that have similar expression patterns. The similarity of the expression data is determined using a novel measure, the slope matrix. The slope matrix introduces a normalized method for the comparison of expression levels throughout a proteome. The algorithm is tested using real microarray gene expression data. Their functions are characterized using gene ontology annotations. The results of the case study indicate the protein function prediction algorithm developed is comparable to the prediction algorithms that are based on the annotations of homologous proteins. -

Role of Mitochondrial Ribosomal Protein S18-2 in Cancerogenesis and in Regulation of Stemness and Differentiation
From THE DEPARTMENT OF MICROBIOLOGY TUMOR AND CELL BIOLOGY (MTC) Karolinska Institutet, Stockholm, Sweden ROLE OF MITOCHONDRIAL RIBOSOMAL PROTEIN S18-2 IN CANCEROGENESIS AND IN REGULATION OF STEMNESS AND DIFFERENTIATION Muhammad Mushtaq Stockholm 2017 All previously published papers were reproduced with permission from the publisher. Published by Karolinska Institutet. Printed by E-Print AB 2017 © Muhammad Mushtaq, 2017 ISBN 978-91-7676-697-2 Role of Mitochondrial Ribosomal Protein S18-2 in Cancerogenesis and in Regulation of Stemness and Differentiation THESIS FOR DOCTORAL DEGREE (Ph.D.) By Muhammad Mushtaq Principal Supervisor: Faculty Opponent: Associate Professor Elena Kashuba Professor Pramod Kumar Srivastava Karolinska Institutet University of Connecticut Department of Microbiology Tumor and Cell Center for Immunotherapy of Cancer and Biology (MTC) Infectious Diseases Co-supervisor(s): Examination Board: Professor Sonia Lain Professor Ola Söderberg Karolinska Institutet Uppsala University Department of Microbiology Tumor and Cell Department of Immunology, Genetics and Biology (MTC) Pathology (IGP) Professor George Klein Professor Boris Zhivotovsky Karolinska Institutet Karolinska Institutet Department of Microbiology Tumor and Cell Institute of Environmental Medicine (IMM) Biology (MTC) Professor Lars-Gunnar Larsson Karolinska Institutet Department of Microbiology Tumor and Cell Biology (MTC) Dedicated to my parents ABSTRACT Mitochondria carry their own ribosomes (mitoribosomes) for the translation of mRNA encoded by mitochondrial DNA. The architecture of mitoribosomes is mainly composed of mitochondrial ribosomal proteins (MRPs), which are encoded by nuclear genomic DNA. Emerging experimental evidences reveal that several MRPs are multifunctional and they exhibit important extra-mitochondrial functions, such as involvement in apoptosis, protein biosynthesis and signal transduction. Dysregulations of the MRPs are associated with severe pathological conditions, including cancer. -

1 AGING Supplementary Table 2
SUPPLEMENTARY TABLES Supplementary Table 1. Details of the eight domain chains of KIAA0101. Serial IDENTITY MAX IN COMP- INTERFACE ID POSITION RESOLUTION EXPERIMENT TYPE number START STOP SCORE IDENTITY LEX WITH CAVITY A 4D2G_D 52 - 69 52 69 100 100 2.65 Å PCNA X-RAY DIFFRACTION √ B 4D2G_E 52 - 69 52 69 100 100 2.65 Å PCNA X-RAY DIFFRACTION √ C 6EHT_D 52 - 71 52 71 100 100 3.2Å PCNA X-RAY DIFFRACTION √ D 6EHT_E 52 - 71 52 71 100 100 3.2Å PCNA X-RAY DIFFRACTION √ E 6GWS_D 41-72 41 72 100 100 3.2Å PCNA X-RAY DIFFRACTION √ F 6GWS_E 41-72 41 72 100 100 2.9Å PCNA X-RAY DIFFRACTION √ G 6GWS_F 41-72 41 72 100 100 2.9Å PCNA X-RAY DIFFRACTION √ H 6IIW_B 2-11 2 11 100 100 1.699Å UHRF1 X-RAY DIFFRACTION √ www.aging-us.com 1 AGING Supplementary Table 2. Significantly enriched gene ontology (GO) annotations (cellular components) of KIAA0101 in lung adenocarcinoma (LinkedOmics). Leading Description FDR Leading Edge Gene EdgeNum RAD51, SPC25, CCNB1, BIRC5, NCAPG, ZWINT, MAD2L1, SKA3, NUF2, BUB1B, CENPA, SKA1, AURKB, NEK2, CENPW, HJURP, NDC80, CDCA5, NCAPH, BUB1, ZWILCH, CENPK, KIF2C, AURKA, CENPN, TOP2A, CENPM, PLK1, ERCC6L, CDT1, CHEK1, SPAG5, CENPH, condensed 66 0 SPC24, NUP37, BLM, CENPE, BUB3, CDK2, FANCD2, CENPO, CENPF, BRCA1, DSN1, chromosome MKI67, NCAPG2, H2AFX, HMGB2, SUV39H1, CBX3, TUBG1, KNTC1, PPP1CC, SMC2, BANF1, NCAPD2, SKA2, NUP107, BRCA2, NUP85, ITGB3BP, SYCE2, TOPBP1, DMC1, SMC4, INCENP. RAD51, OIP5, CDK1, SPC25, CCNB1, BIRC5, NCAPG, ZWINT, MAD2L1, SKA3, NUF2, BUB1B, CENPA, SKA1, AURKB, NEK2, ESCO2, CENPW, HJURP, TTK, NDC80, CDCA5, BUB1, ZWILCH, CENPK, KIF2C, AURKA, DSCC1, CENPN, CDCA8, CENPM, PLK1, MCM6, ERCC6L, CDT1, HELLS, CHEK1, SPAG5, CENPH, PCNA, SPC24, CENPI, NUP37, FEN1, chromosomal 94 0 CENPL, BLM, KIF18A, CENPE, MCM4, BUB3, SUV39H2, MCM2, CDK2, PIF1, DNA2, region CENPO, CENPF, CHEK2, DSN1, H2AFX, MCM7, SUV39H1, MTBP, CBX3, RECQL4, KNTC1, PPP1CC, CENPP, CENPQ, PTGES3, NCAPD2, DYNLL1, SKA2, HAT1, NUP107, MCM5, MCM3, MSH2, BRCA2, NUP85, SSB, ITGB3BP, DMC1, INCENP, THOC3, XPO1, APEX1, XRCC5, KIF22, DCLRE1A, SEH1L, XRCC3, NSMCE2, RAD21. -
![MRPL10 Mouse Monoclonal Antibody [Clone ID: OTI2B4] Product Data](https://docslib.b-cdn.net/cover/0766/mrpl10-mouse-monoclonal-antibody-clone-id-oti2b4-product-data-610766.webp)
MRPL10 Mouse Monoclonal Antibody [Clone ID: OTI2B4] Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TA808643 MRPL10 Mouse Monoclonal Antibody [Clone ID: OTI2B4] Product data: Product Type: Primary Antibodies Clone Name: OTI2B4 Applications: IHC, WB Recommended Dilution: WB 1:2000, IHC 1:150 Reactivity: Human Host: Mouse Isotype: IgG2b Clonality: Monoclonal Immunogen: Full length human recombinant protein of human MRPL10(NP_660298) produced in E.coli. Formulation: PBS (PH 7.3) containing 1% BSA, 50% glycerol and 0.02% sodium azide. Concentration: 1 mg/ml Purification: Purified from mouse ascites fluids or tissue culture supernatant by affinity chromatography (protein A/G) Conjugation: Unconjugated Storage: Store at -20°C as received. Stability: Stable for 12 months from date of receipt. Predicted Protein Size: 26.3 kDa Gene Name: mitochondrial ribosomal protein L10 Database Link: NP_660298 Entrez Gene 124995 Human Q7Z7H8 This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 5 MRPL10 Mouse Monoclonal Antibody [Clone ID: OTI2B4] – TA808643 Background: Mammalian mitochondrial ribosomal proteins are encoded by nuclear genes and help in protein synthesis within the mitochondrion. Mitochondrial ribosomes (mitoribosomes) consist of a small 28S subunit and a large 39S subunit. They have an estimated 75% protein to rRNA composition compared to prokaryotic ribosomes, where this ratio is reversed. Another difference between mammalian mitoribosomes and prokaryotic ribosomes is that the latter contain a 5S rRNA. -

Mrpl10 and Tbp Are Suitable Reference Genes for Peripheral Nerve Crush Injury
International Journal of Molecular Sciences Article Mrpl10 and Tbp Are Suitable Reference Genes for Peripheral Nerve Crush Injury Yaxian Wang 1,2,†, Qianqian Shan 3,†, Yali Meng 2, Jiacheng Pan 2 and Sheng Yi 2,* 1 School of Biology and Basic Medical Sciences, Soochow University, Suzhou 215123, China; [email protected] 2 Key Laboratory of Neuroregeneration of Jiangsu and Ministry of Education, Co-Innovation Center of Neuroregeneration, Nantong University, Nantong 226001, China; [email protected] (Y.M.); [email protected] (J.P.) 3 Department of Radiotherapy and Oncology, The Affiliated Hospital of Nantong University, Nantong 226001, China; [email protected] * Correspondence: [email protected]; Tel./Fax: +86-513-8551-1585 † These authors contributed equally to this work. Academic Editor: Irmgard Tegeder Received: 21 December 2016; Accepted: 23 January 2017; Published: 27 January 2017 Abstract: Peripheral nerve injury triggers the dysregulation of a large number of genes at multiple sites, including neurons, peripheral nerve stump, and the target organ. Housekeeping genes were frequently used as reference genes to normalize the expression values of target genes. Suitable selection of housekeeping genes that are stably expressed after nerve injury minimizes bias elicited by reference genes and thus helps to better and more sensitively reflect gene expression changes. However, many housekeeping genes have been used as reference genes without testing the expression patterns of themselves. In the current study, we calculated the expression -

Using Edsurvey to Analyze NCES Data: an Illustration of Analyzing NAEP Primer
Using EdSurvey to Analyze NCES Data: An Illustration of Analyzing NAEP Primer Developed by Michael Lee, Paul Bailey, Ahmad Emad, Ting Zhang, Trang Nguyen, and Jiao Yu*† February 21, 2020 Overview of the EdSurvey Package National Assessment of Educational Progress (NAEP) datasets from the National Center for Education Statistics (NCES) require special statistical methods to analyze. Because of their scope and complexity, the EdSurvey package gives users functions to perform analyses that account for both complex sample survey designs and the use of plausible values. The EdSurvey package also seamlessly takes advantage of the LaF package to read in data only when required for an analysis. Users with computers that have insuÿcient memory to read in entire NAEP datasets can still do analyses without having to write special code to read in just the appropriate variables. This situation is addressed directly in the EdSurvey package—behind the scenes and without any special tuning by the user. Vignette Outline This vignette will describe the basics of using the EdSurvey package for analyzing NAEP data as follows. • Notes – Additional resources – Vignette notation – Software requirements • Setting up the environment for analyzing NCES data – Installing and loading EdSurvey – Philosophy of Conducting Analyses Using the EdSurvey Package – Downloading data – Reading in data – Getting to know the data format – Removing special values • Explore Variable Distributions with summary2 • Subsetting the data • Retrieving data for further manipulation with getData *This publication was prepared for NCES under Contract No. ED-IES-12-D-0002 with the American Institutes for Research. Mention of trade names, commercial products, or organizations does not imply endorsement by the U.S. -

Open FINAL GRAD SCHOOL.Pdf
The Pennsylvania State University The Graduate School Eberly College of Science PHOSPHOPROTEOMIC ANALYSIS OF RIBOSOMAL PROTEINS: IMPLICATIONS IN TRANSLATION AND APOPTOSIS A Dissertation in Biochemistry, Microbiology, and Molecular Biology by Jennifer Lynn Miller © 2009 Jennifer Lynn Miller Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy May 2009 The dissertation of Jennifer Lynn Miller was reviewed and approved* by the following: Emine C. Koc Assistant Professor Biochemistry and Molecular Biology Dissertation Advisor Chair of Committee Robert A. Schlegel Professor of Biochemistry and Molecular Biology Wendy Hanna-Rose Assistant Professor Biochemistry and Molecular Biology Ming Tien Professor of Biochemistry Erin D. Sheets Assistant Professor of Chemistry Richard J. Frisque Professor of Molecular Virology Head of the Department of Biochemistry and Molecular Biology *Signatures are on file in the Graduate School. ABSTRACT Mammalian mitochondrial ribosomes synthesize thirteen proteins that are essential for oxidative phosphorylation. Besides having a major role in ATP synthesis, mitochondria also contribute to biochemical processes coordinating apoptosis, mitochondrial diseases, and aging in eukaryotic cells. This unique class of ribosomes is protein-rich and distinct from cytoplasmic ribosomes. However, mitochondrial ribosomes (55S) share a significant homology to bacterial ribosomes (70S), particularly in size, the general mechanism of translation, and ribosomal protein content. Due to the overall resemblance between the two systems and the earlier reports of post-translational modifications, we investigated how phosphorylation of ribosomal proteins from bacteria and mitochondria regulates translation and other acquired roles. Identification of twenty- four phosphorylated 70S and 55S ribosomal proteins as well as the potential endogenous kinase was achieved using 2D-gel electrophoresis and tandem mass spectrometry. -
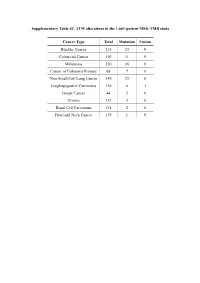
Supplementary Table S1. ATM Alterations in the 1,661-Patient MSK-TMB Study
Supplementary Table S1. ATM alterations in the 1,661-patient MSK-TMB study Cancer Type Total Mutation Fusion Bladder Cancer 215 23 0 Colorectal Cancer 110 11 0 Melanoma 320 26 0 Cancer of Unknown Primary 88 7 0 Non-Small Cell Lung Cancer 350 23 0 Esophagogastric Carcinoma 126 6 1 Breast Cancer 44 2 0 Glioma 117 3 0 Renal Cell Carcinoma 151 2 0 Head and Neck Cancer 139 1 0 Supplementary Table S2. ATM alternations in the 10,945-patient MSK-IMPACT study Cancer Type Total Mutation Deep deletion Amplification Multi-alterations Fusion Small Bowl Cancer 35 7 0 0 0 0 Skin Cancer, Non-Melaoma 148 19 1 0 0 0 Bladder Cancer 423 45 2 0 0 0 Endometrial Cancer 218 24 0 0 0 0 Hepatobiliary Cancer 355 27 0 0 0 0 Colorectal Cancer 1007 75 0 0 0 1 Mature B-Cell Neoplasms 134 8 0 0 2 0 Non-Small Cell Lung Cancer 1668 120 0 2 1 0 Melanoma 365 23 0 0 0 0 Appendiceal Cancer 79 4 0 0 0 0 Small Cell Lung Cancer 82 4 0 0 0 0 Prostate Cancer 717 25 7 1 0 0 Histiocytosis 22 1 0 0 0 0 Salivary Gland Cancer 114 5 0 0 0 0 Thyroid Cancer 231 10 0 0 0 0 Cancer of Unknown Primary 186 8 0 0 0 0 Breast Cancer 1324 48 3 2 0 0 Adrenocortical Carcinoma 25 1 0 0 0 0 Mature T and NK Neoplasms 29 1 0 0 0 0 Renal Cell Carcinoma 361 12 0 0 0 0 Head and Neck Cancer 186 14 0 0 0 1 Pancreatic Cancer 502 69 7 2 0 0 Glioma 553 14 1 0 0 0 Soft Tissue Sarcoma 443 13 1 0 0 0 Esophagogastric Cancer 341 8 1 0 0 0 Peripheral Nervous System 80 0 2 0 0 0 Germ Cell Tumor 288 2 0 1 0 1 Ovarian Can 244 2 1 0 0 0 Uterine Sarcoma 93 1 0 0 0 0 Mesothelioma 107 1 0 0 0 0 Bone Cancer 134 1 0 0 0 0 Gastrointestinal Stromal Ca 137 0 0 0 0 1 Supplementary Table S3. -

Supplementary Dataset S2
mitochondrial translational termination MRPL28 MRPS26 6 MRPS21 PTCD3 MTRF1L 4 MRPL50 MRPS18A MRPS17 2 MRPL20 MRPL52 0 MRPL17 MRPS33 MRPS15 −2 MRPL45 MRPL30 MRPS27 AURKAIP1 MRPL18 MRPL3 MRPS6 MRPS18B MRPL41 MRPS2 MRPL34 GADD45GIP1 ERAL1 MRPL37 MRPS10 MRPL42 MRPL19 MRPS35 MRPL9 MRPL24 MRPS5 MRPL44 MRPS23 MRPS25 ITB ITB ITB ITB ICa ICr ITL original ICr ICa ITL ICa ITL original ICr ITL ICr ICa mitochondrial translational elongation MRPL28 MRPS26 6 MRPS21 PTCD3 MRPS18A 4 MRPS17 MRPL20 2 MRPS15 MRPL45 MRPL52 0 MRPS33 MRPL30 −2 MRPS27 AURKAIP1 MRPS10 MRPL42 MRPL19 MRPL18 MRPL3 MRPS6 MRPL24 MRPS35 MRPL9 MRPS18B MRPL41 MRPS2 MRPL34 MRPS5 MRPL44 MRPS23 MRPS25 MRPL50 MRPL17 GADD45GIP1 ERAL1 MRPL37 ITB ITB ITB ITB ICa ICr original ICr ITL ICa ITL ICa ITL original ICr ITL ICr ICa translational termination MRPL28 MRPS26 6 MRPS21 PTCD3 C12orf65 4 MTRF1L MRPL50 MRPS18A 2 MRPS17 MRPL20 0 MRPL52 MRPL17 MRPS33 −2 MRPS15 MRPL45 MRPL30 MRPS27 AURKAIP1 MRPL18 MRPL3 MRPS6 MRPS18B MRPL41 MRPS2 MRPL34 GADD45GIP1 ERAL1 MRPL37 MRPS10 MRPL42 MRPL19 MRPS35 MRPL9 MRPL24 MRPS5 MRPL44 MRPS23 MRPS25 ITB ITB ITB ITB ICa ICr original ICr ITL ICa ITL ICa ITL original ICr ITL ICr ICa translational elongation DIO2 MRPS18B MRPL41 6 MRPS2 MRPL34 GADD45GIP1 4 ERAL1 MRPL37 2 MRPS10 MRPL42 MRPL19 0 MRPL30 MRPS27 AURKAIP1 −2 MRPL18 MRPL3 MRPS6 MRPS35 MRPL9 EEF2K MRPL50 MRPS5 MRPL44 MRPS23 MRPS25 MRPL24 MRPS33 MRPL52 EIF5A2 MRPL17 SECISBP2 MRPS15 MRPL45 MRPS18A MRPS17 MRPL20 MRPL28 MRPS26 MRPS21 PTCD3 ITB ITB ITB ITB ICa ICr ICr ITL original ITL ICa ICa ITL ICr ICr ICa original -

The Pennsylvania State University the Graduate School Eberly
The Pennsylvania State University The Graduate School Eberly College of Science REGULATION OF MITOCHONDRIAL TRANSLATION AND OXIDATIVE PHOSPHORYLATION THROUGH REVERSIBLE ACETYLATION A Dissertation in Biochemistry, Microbiology and Molecular Biology by Hüseyin Çimen 2012 Hüseyin Çimen Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy August 2012 The Dissertation of Hüseyin Çimen was reviewed and approved* by the following: Emine C. Koc Assistant Professor of Biochemistry and Molecular Biology Dissertation Co-adviser Co-chair of Committee Hasan Koc Assistant Professor of Natural Sciences Dissertation Co-adviser Co-chair of Committee Craig E. Cameron Paul Berg Professor of Biochemistry and Molecular Biology Associate Department Head for Research and Graduate Education Joseph C. Reese Professor of Biochemistry and Molecular Biology Teh-hui Kao Professor of Biochemistry and Molecular Biology Tae-Hee Lee Assistant Professor of Chemistry and the Huck Institute of the Life Sciences Craig E. Cameron Paul Berg Professor of Biochemistry and Molecular Biology Associate Department Head of the Department of Biochemistry and Molecular Biology iii ABSTRACT In a eukaryotic cell, mitochondria provide energy in the form of ATP through oxidative phosphorylation (OXPHOS), which consists of five electron transport chain complexes embedded in the inner membrane of mitochondria. Human mitochondria have their own genome and transcription/translation system to synthesize mitochondrially encoded thirteen proteins of respiratory chain complexes. We investigated how acetylation of ribosomal proteins regulates translation and energy production in mitochondria since reversible acetylation of mitochondrial proteins was found to be critical for maintaining energy homeostasis. We identified mitochondrial ribosomal protein L10 (MRPL10) as the major acetylated ribosomal protein in mammalian mitochondria with two-dimensional gel electrophoresis followed by tandem mass spectrometry and immunoblotting analyses. -

C6orf203 Controls OXPHOS Function Through Modulation of Mitochondrial Protein Biosynthesis
bioRxiv preprint doi: https://doi.org/10.1101/704403; this version posted July 17, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. C6orf203 controls OXPHOS function through modulation of mitochondrial protein biosynthesis number of characters excluding Materials and Methods: 40,651 Sara Palacios-Zambrano1,2, Luis Vázquez-Fonseca1,2, Cristina González-Páramos1,2, Laura Mamblona1,2, Laura Sánchez-Caballero3, Leo Nijtmans3, Rafael Garesse1,2 and Miguel Angel Fernández-Moreno1,2,* 1 Departamento de Bioquímica, Instituto de Investigaciones Biomédicas “Alberto Sols” UAM CSIC and Centro de Investigación Biomédica en Red en Enfermedades Raras (CIBERER). Facultad de Medicina, Universidad Autónoma de Madrid. Madrid 28029, Spain. 2 Instituto de Investigación Sanitaria Hospital 12 de Octubre (imas12), Madrid 28041, Spain. 3 Department of Pediatrics, Radboud Center for Mitochondrial Medicine, Radboud University Medical Center, Nijmegen, The Netherlands. * To whom correspondence should be addressed. Tel:+34 91 497 31 29; Email: [email protected] Running title “C6orf203 controls mt-proteins synthesis” bioRxiv preprint doi: https://doi.org/10.1101/704403; this version posted July 17, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. ABSTRACT Mitochondria are essential organelles present in the vast majority of eukaryotic cells. Their central function is to produce cellular energy through the OXPHOS system, and functional alterations provoke so-called mitochondrial OXPHOS diseases. It is estimated that several hundred mitochondrial proteins have unknown functions. Very recently, C6orf203 was described to participate in mitochondrial transcription under induced mitochondrial DNA depletion stress conditions. -

Human MRPS24 ORF Mammalian Expression Plasmid, N-Myc Tag
Human MRPS24 ORF mammalian expression plasmid, N-Myc tag Catalog Number: HG16377-NM General Information Plasmid Resuspension protocol Gene : mitochondrial ribosomal protein S24 1. Centrifuge at 5,000×g for 5 min. Official Symbol : MRPS24 2. Carefully open the tube and add 100 l of sterile water to Synonym : S24mt, bMRP47, HSPC335, MRP-S24, dissolve the DNA. bMRP-47 3. Close the tube and incubate for 10 minutes at room Source : Human temperature. cDNA Size: 504bp 4. Briefly vortex the tube and then do a quick spin to RefSeq : NM_032014.2 concentrate the liquid at the bottom. Speed is less than Description 5000×g. Lot : Please refer to the label on the tube 5. Store the plasmid at -20 ℃. Vector : pCMV3-N-Myc Shipping carrier : Each tube contains approximately 10 μg of lyophilized plasmid. The plasmid is ready for: Storage : • Restriction enzyme digestion The lyophilized plasmid can be stored at ambient temperature • PCR amplification for three months. • E. coli transformation Quality control : • DNA sequencing The plasmid is confirmed by full-length sequencing with primers in the sequencing primer list. E.coli strains for transformation (recommended Sequencing primer list : but not limited) pCMV3-F: 5’ CAGGTGTCCACTCCCAGGTCCAAG 3’ Most commercially available competent cells are appropriate for pcDNA3-R : 5’ GGCAACTAGAAGGCACAGTCGAGG 3’ the plasmid, e.g. TOP10, DH5α and TOP10F´. Or Forward T7 : 5’ TAATACGACTCACTATAGGG 3’ ReverseBGH : 5’ TAGAAGGCACAGTCGAGG 3’ pCMV3-F and pcDNA3-R are designed by Sino Biological Inc. Customers can order the primer pair from any oligonucleotide supplier. Manufactured By Sino Biological Inc., FOR RESEARCH USE ONLY. NOT FOR USE IN HUMANS.