Mrpl10 and Tbp Are Suitable Reference Genes for Peripheral Nerve Crush Injury
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Role of Mitochondrial Ribosomal Protein S18-2 in Cancerogenesis and in Regulation of Stemness and Differentiation
From THE DEPARTMENT OF MICROBIOLOGY TUMOR AND CELL BIOLOGY (MTC) Karolinska Institutet, Stockholm, Sweden ROLE OF MITOCHONDRIAL RIBOSOMAL PROTEIN S18-2 IN CANCEROGENESIS AND IN REGULATION OF STEMNESS AND DIFFERENTIATION Muhammad Mushtaq Stockholm 2017 All previously published papers were reproduced with permission from the publisher. Published by Karolinska Institutet. Printed by E-Print AB 2017 © Muhammad Mushtaq, 2017 ISBN 978-91-7676-697-2 Role of Mitochondrial Ribosomal Protein S18-2 in Cancerogenesis and in Regulation of Stemness and Differentiation THESIS FOR DOCTORAL DEGREE (Ph.D.) By Muhammad Mushtaq Principal Supervisor: Faculty Opponent: Associate Professor Elena Kashuba Professor Pramod Kumar Srivastava Karolinska Institutet University of Connecticut Department of Microbiology Tumor and Cell Center for Immunotherapy of Cancer and Biology (MTC) Infectious Diseases Co-supervisor(s): Examination Board: Professor Sonia Lain Professor Ola Söderberg Karolinska Institutet Uppsala University Department of Microbiology Tumor and Cell Department of Immunology, Genetics and Biology (MTC) Pathology (IGP) Professor George Klein Professor Boris Zhivotovsky Karolinska Institutet Karolinska Institutet Department of Microbiology Tumor and Cell Institute of Environmental Medicine (IMM) Biology (MTC) Professor Lars-Gunnar Larsson Karolinska Institutet Department of Microbiology Tumor and Cell Biology (MTC) Dedicated to my parents ABSTRACT Mitochondria carry their own ribosomes (mitoribosomes) for the translation of mRNA encoded by mitochondrial DNA. The architecture of mitoribosomes is mainly composed of mitochondrial ribosomal proteins (MRPs), which are encoded by nuclear genomic DNA. Emerging experimental evidences reveal that several MRPs are multifunctional and they exhibit important extra-mitochondrial functions, such as involvement in apoptosis, protein biosynthesis and signal transduction. Dysregulations of the MRPs are associated with severe pathological conditions, including cancer. -

1 AGING Supplementary Table 2
SUPPLEMENTARY TABLES Supplementary Table 1. Details of the eight domain chains of KIAA0101. Serial IDENTITY MAX IN COMP- INTERFACE ID POSITION RESOLUTION EXPERIMENT TYPE number START STOP SCORE IDENTITY LEX WITH CAVITY A 4D2G_D 52 - 69 52 69 100 100 2.65 Å PCNA X-RAY DIFFRACTION √ B 4D2G_E 52 - 69 52 69 100 100 2.65 Å PCNA X-RAY DIFFRACTION √ C 6EHT_D 52 - 71 52 71 100 100 3.2Å PCNA X-RAY DIFFRACTION √ D 6EHT_E 52 - 71 52 71 100 100 3.2Å PCNA X-RAY DIFFRACTION √ E 6GWS_D 41-72 41 72 100 100 3.2Å PCNA X-RAY DIFFRACTION √ F 6GWS_E 41-72 41 72 100 100 2.9Å PCNA X-RAY DIFFRACTION √ G 6GWS_F 41-72 41 72 100 100 2.9Å PCNA X-RAY DIFFRACTION √ H 6IIW_B 2-11 2 11 100 100 1.699Å UHRF1 X-RAY DIFFRACTION √ www.aging-us.com 1 AGING Supplementary Table 2. Significantly enriched gene ontology (GO) annotations (cellular components) of KIAA0101 in lung adenocarcinoma (LinkedOmics). Leading Description FDR Leading Edge Gene EdgeNum RAD51, SPC25, CCNB1, BIRC5, NCAPG, ZWINT, MAD2L1, SKA3, NUF2, BUB1B, CENPA, SKA1, AURKB, NEK2, CENPW, HJURP, NDC80, CDCA5, NCAPH, BUB1, ZWILCH, CENPK, KIF2C, AURKA, CENPN, TOP2A, CENPM, PLK1, ERCC6L, CDT1, CHEK1, SPAG5, CENPH, condensed 66 0 SPC24, NUP37, BLM, CENPE, BUB3, CDK2, FANCD2, CENPO, CENPF, BRCA1, DSN1, chromosome MKI67, NCAPG2, H2AFX, HMGB2, SUV39H1, CBX3, TUBG1, KNTC1, PPP1CC, SMC2, BANF1, NCAPD2, SKA2, NUP107, BRCA2, NUP85, ITGB3BP, SYCE2, TOPBP1, DMC1, SMC4, INCENP. RAD51, OIP5, CDK1, SPC25, CCNB1, BIRC5, NCAPG, ZWINT, MAD2L1, SKA3, NUF2, BUB1B, CENPA, SKA1, AURKB, NEK2, ESCO2, CENPW, HJURP, TTK, NDC80, CDCA5, BUB1, ZWILCH, CENPK, KIF2C, AURKA, DSCC1, CENPN, CDCA8, CENPM, PLK1, MCM6, ERCC6L, CDT1, HELLS, CHEK1, SPAG5, CENPH, PCNA, SPC24, CENPI, NUP37, FEN1, chromosomal 94 0 CENPL, BLM, KIF18A, CENPE, MCM4, BUB3, SUV39H2, MCM2, CDK2, PIF1, DNA2, region CENPO, CENPF, CHEK2, DSN1, H2AFX, MCM7, SUV39H1, MTBP, CBX3, RECQL4, KNTC1, PPP1CC, CENPP, CENPQ, PTGES3, NCAPD2, DYNLL1, SKA2, HAT1, NUP107, MCM5, MCM3, MSH2, BRCA2, NUP85, SSB, ITGB3BP, DMC1, INCENP, THOC3, XPO1, APEX1, XRCC5, KIF22, DCLRE1A, SEH1L, XRCC3, NSMCE2, RAD21. -
![MRPL10 Mouse Monoclonal Antibody [Clone ID: OTI2B4] Product Data](https://docslib.b-cdn.net/cover/0766/mrpl10-mouse-monoclonal-antibody-clone-id-oti2b4-product-data-610766.webp)
MRPL10 Mouse Monoclonal Antibody [Clone ID: OTI2B4] Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TA808643 MRPL10 Mouse Monoclonal Antibody [Clone ID: OTI2B4] Product data: Product Type: Primary Antibodies Clone Name: OTI2B4 Applications: IHC, WB Recommended Dilution: WB 1:2000, IHC 1:150 Reactivity: Human Host: Mouse Isotype: IgG2b Clonality: Monoclonal Immunogen: Full length human recombinant protein of human MRPL10(NP_660298) produced in E.coli. Formulation: PBS (PH 7.3) containing 1% BSA, 50% glycerol and 0.02% sodium azide. Concentration: 1 mg/ml Purification: Purified from mouse ascites fluids or tissue culture supernatant by affinity chromatography (protein A/G) Conjugation: Unconjugated Storage: Store at -20°C as received. Stability: Stable for 12 months from date of receipt. Predicted Protein Size: 26.3 kDa Gene Name: mitochondrial ribosomal protein L10 Database Link: NP_660298 Entrez Gene 124995 Human Q7Z7H8 This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 5 MRPL10 Mouse Monoclonal Antibody [Clone ID: OTI2B4] – TA808643 Background: Mammalian mitochondrial ribosomal proteins are encoded by nuclear genes and help in protein synthesis within the mitochondrion. Mitochondrial ribosomes (mitoribosomes) consist of a small 28S subunit and a large 39S subunit. They have an estimated 75% protein to rRNA composition compared to prokaryotic ribosomes, where this ratio is reversed. Another difference between mammalian mitoribosomes and prokaryotic ribosomes is that the latter contain a 5S rRNA. -

The Pennsylvania State University the Graduate School Eberly
The Pennsylvania State University The Graduate School Eberly College of Science REGULATION OF MITOCHONDRIAL TRANSLATION AND OXIDATIVE PHOSPHORYLATION THROUGH REVERSIBLE ACETYLATION A Dissertation in Biochemistry, Microbiology and Molecular Biology by Hüseyin Çimen 2012 Hüseyin Çimen Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy August 2012 The Dissertation of Hüseyin Çimen was reviewed and approved* by the following: Emine C. Koc Assistant Professor of Biochemistry and Molecular Biology Dissertation Co-adviser Co-chair of Committee Hasan Koc Assistant Professor of Natural Sciences Dissertation Co-adviser Co-chair of Committee Craig E. Cameron Paul Berg Professor of Biochemistry and Molecular Biology Associate Department Head for Research and Graduate Education Joseph C. Reese Professor of Biochemistry and Molecular Biology Teh-hui Kao Professor of Biochemistry and Molecular Biology Tae-Hee Lee Assistant Professor of Chemistry and the Huck Institute of the Life Sciences Craig E. Cameron Paul Berg Professor of Biochemistry and Molecular Biology Associate Department Head of the Department of Biochemistry and Molecular Biology iii ABSTRACT In a eukaryotic cell, mitochondria provide energy in the form of ATP through oxidative phosphorylation (OXPHOS), which consists of five electron transport chain complexes embedded in the inner membrane of mitochondria. Human mitochondria have their own genome and transcription/translation system to synthesize mitochondrially encoded thirteen proteins of respiratory chain complexes. We investigated how acetylation of ribosomal proteins regulates translation and energy production in mitochondria since reversible acetylation of mitochondrial proteins was found to be critical for maintaining energy homeostasis. We identified mitochondrial ribosomal protein L10 (MRPL10) as the major acetylated ribosomal protein in mammalian mitochondria with two-dimensional gel electrophoresis followed by tandem mass spectrometry and immunoblotting analyses. -

Transcriptomic and Proteomic Landscape of Mitochondrial
TOOLS AND RESOURCES Transcriptomic and proteomic landscape of mitochondrial dysfunction reveals secondary coenzyme Q deficiency in mammals Inge Ku¨ hl1,2†*, Maria Miranda1†, Ilian Atanassov3, Irina Kuznetsova4,5, Yvonne Hinze3, Arnaud Mourier6, Aleksandra Filipovska4,5, Nils-Go¨ ran Larsson1,7* 1Department of Mitochondrial Biology, Max Planck Institute for Biology of Ageing, Cologne, Germany; 2Department of Cell Biology, Institute of Integrative Biology of the Cell (I2BC) UMR9198, CEA, CNRS, Univ. Paris-Sud, Universite´ Paris-Saclay, Gif- sur-Yvette, France; 3Proteomics Core Facility, Max Planck Institute for Biology of Ageing, Cologne, Germany; 4Harry Perkins Institute of Medical Research, The University of Western Australia, Nedlands, Australia; 5School of Molecular Sciences, The University of Western Australia, Crawley, Australia; 6The Centre National de la Recherche Scientifique, Institut de Biochimie et Ge´ne´tique Cellulaires, Universite´ de Bordeaux, Bordeaux, France; 7Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Stockholm, Sweden Abstract Dysfunction of the oxidative phosphorylation (OXPHOS) system is a major cause of human disease and the cellular consequences are highly complex. Here, we present comparative *For correspondence: analyses of mitochondrial proteomes, cellular transcriptomes and targeted metabolomics of five [email protected] knockout mouse strains deficient in essential factors required for mitochondrial DNA gene (IKu¨ ); expression, leading to OXPHOS dysfunction. Moreover, -
Additional Tables.Xlsx
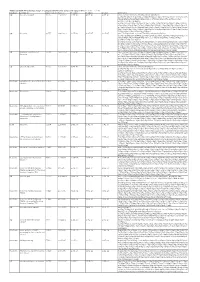
Additional Table 5 Enriched pathways of upregulated DEGs after spinal cord injury in WT (P < 0.05, q < 0.05) Database Description Ratio of DEGs Ratio of P -value P adjust q value gene name GO ribosomal subunit 87/755 214/12262 1.03E-49 4.61E-47 4.20E-47 Gm6576/Gm10036/Gm5786/Mrps28/Gm9493/Rpl13- ps3/Rpl7l1/Mrpl10/Mrpl44/Rps20/Mrpl51/Mrps11/Mpv17l2/Mrps7/Gm10020/Gm10073 /Mrpl2/Mrpl52/Rpl22/Mrpl4/Mrps24/Zcchc17/Rpl28/Mrpl34/Rpl29/Mrps36/Rpl9- ps6/Hba-a2/Hba-a1/Rpl23a- ps3/Rpl15/Rack1/Mrps18a/Mrpl28/Rpl10a/Mrpl12/Rpl18/Rpl35a/Mrpl41/Mrps12/Rpl36 a/Rps9/Rps18/Rps5/Rps27a/Rps24/Rplp2/Rpl14/Mrpl30/Rps6/Rpl6/Rps3/Rps4x/Rpsa/R pl7/Rpl27a/Rps23/Rps10/Mrps21/Rpl36/Rpl30/Rps17/Mrpl42/Rpl35/Rpl27/Rplp0/Rps28 /Rpl31/Rpl39/Rps13/Rpl13/Rps11/Rpl8/Rps3a1/Fau/Rpl9/Rpl3/Rpl26/Rps15/Rpl13a/Uba 52/Rpl24/Rpl19/Rps14/Rpl4/Rps27/Rplp1 GO ribosome 93/755 248/12262 1.67E-49 4.61E-47 4.20E-47 Gm6576/Pnpt1/Gm10036/Gm5786/Mrps28/Gm9493/Rpl13- ps3/Rpl7l1/Mrpl10/Mrpl44/Rps20/Mrpl51/Mrps11/Mpv17l2/Mrps7/Gm10020/Gm10073 /Mrpl2/Mrpl52/Rpl22/Mrpl4/Mrps24/Zcchc17/Rpl28/Mrpl34/Rpl29/Mrps36/Rpl9- ps6/Hba-a2/Hba-a1/Rpl23a- ps3/Rpl15/Rack1/Eif3h/Mrps18a/Mrpl28/Mrps33/Rpl10a/Mrpl12/Rpl18/Rpl35a/Btf3/Mrp l41/Mrps12/Rpl36a/Rps9/Rps18/Rps5/Rps27a/Rps24/Rplp2/Rpl14/Mrpl30/Rps6/Rpl6/R ps3/Rps4x/Rpsa/Rpl7/Rpl27a/Rps23/Rps10/Mrps21/Rpl36/Rpl30/Rps17/Mrpl42/Rpl35/R pl27/Rplp0/Rps28/Rpl31/Rpl39/Rps13/Rpl13/Rps11/Rpl8/Rps3a1/Fau/Rpl9/Rpl3/Rpl26/ Rps15/Rpl13a/Uba52/Rpl41/Rpl24/Rpl19/Rps14/Rpl4/Ndufa7/Rps27/Rplp1 GO structural constituent of 76/733 169/11958 1.65E-47 1.16E-44 1.12E-44 Gm6576/Gm10036/Gm5786/Rpl7l1/Mrpl10/Rps20/Mrpl51/Mrps11/Mrps7/Gm10020/G -

Mitochondrial Ribosomal Protein L12 Selectively Associates with Human Mitochondrial RNA Polymerase to Activate Transcription
Mitochondrial Ribosomal Protein L12 selectively associates with human mitochondrial RNA polymerase to activate transcription Yulia V. Surovtsevaa, Timothy E. Shutta, Justin Cotneya, Huseyin Cimenb, Sophia Y. Chena, Emine C. Kocb, and Gerald S. Shadela,c,1 aDepartment of Pathology, Yale University School of Medicine, New Haven, CT 06520; cDepartment of Genetics, Yale University School of Medicine, New Haven, CT 06520; and bDepartment of Biochemistry and Molecular Biology, Pennsylvania State University, University Park, PA 16802 Edited* by Douglas C. Wallace, Children's Hospital of Philadelphia, Philadelphia, PA, and approved August 31, 2011 (received for review June 1, 2011) Basal transcription of human mitochondrial DNA (mtDNA) in vitro Based on in vitro studies, transcription of human mtDNA re- requires the single-subunit, bacteriophage-related RNA polymerase, quires a core two-component system (9): a single-subunit RNA POLRMT, and transcription factor h-mtTFB2. This two-component polymerase, POLRMT (10), and human mitochondrial transcrip- system is activated differentially at mtDNA promoters by human tion factor B2, h-mtTFB2 (or TFB2M) (11, 12). POLRMT, like its mitochondrial transcription factor A (h-mtTFA). Mitochondrial ribo- yeast homolog Rpo41p, is related to the bacteriophage T-odd somal protein L7/L12 (MRPL12) binds directly to POLRMT, but family of RNA polymerases (10, 13), but has a large N-terminal whether it does so in the context of the ribosome or as a “free” pro- extension that contains two pentatricopeptide repeats of unknown tein in the matrix is unknown. Furthermore, existing evidence that function (14). Relevant to this study, h-mtTFB2 is homologous to MRPL12 activates mitochondrial transcription derives from overex- the bacterial KsgA family of rRNA methyltransferases, an ortholog pression studies in cultured cells and transcription experiments using of which was inherited from the original bacterial endosymbiont crude mitochondrial lysates, precluding direct effects of MRPL12 on ancestor of mitochondria (15, 16). -

Cardiomyopathy Is Associated with Ribosomal Protein Gene Haploinsufficiency In
Genetics: Published Articles Ahead of Print, published on September 2, 2011 as 10.1534/genetics.111.131482 Cardiomyopathy is associated with ribosomal protein gene haploinsufficiency in Drosophila melanogaster Michelle E. Casad*, Dennis Abraham§, Il-Man Kim§, Stephan Frangakis*, Brian Dong*, Na Lin§1, Matthew J. Wolf§, and Howard A. Rockman*§** Department of *Cell Biology, §Medicine, and **Molecular Genetics and Microbiology, Duke University Medical Center, Durham, NC 27710 1Current Address: Institute of Molecular Medicine, Peking University, Beijing, China, 100871 1 Copyright 2011. Running Title: Cardiomyopathy in Minute Drosophila Key Words: Drosophila Minute syndrome Cardiomyopathy Address for correspondence: Howard A. Rockman, M.D. Department of Medicine Duke University Medical Center, DUMC 3104 226 CARL Building, Research Drive Durham, NC, 27710 Tel: (919) 668-2520 FAX: (919) 668-2524 Email: [email protected] 2 ABSTRACT The Minute syndrome in Drosophila melanogaster is characterized by delayed development, poor fertility, and short slender bristles. Many Minute loci correspond to disruptions of genes for cytoplasmic ribosomal proteins, and therefore the phenotype has been attributed to alterations in translational processes. Although protein translation is crucial for all cells in an organism, it is unclear why Minute mutations cause effects in specific tissues. To determine whether the heart is sensitive to haploinsufficiency of genes encoding ribosomal proteins, we measured heart function of Minute mutants using optical coherence tomography. We found that cardiomyopathy is associated with the Minute syndrome caused by haploinsufficiency of genes encoding cytoplasmic ribosomal proteins. While mutations of genes encoding non-Minute cytoplasmic ribosomal proteins are homozygous lethal, heterozygous deficiencies spanning these non-Minute genes did not cause a change in cardiac function. -

MRPL10 Polyclonal Antibody
PRODUCT DATA SHEET Bioworld Technology,Inc. MRPL10 polyclonal antibody Catalog: BS65315 Host: Rabbit Reactivity: Human BackGround: term. Avoid freeze-thaw cycles. mitochondrial ribosomal protein L10(MRPL10) Homo Specificity: sapiens Mammalian mitochondrial ribosomal proteins are MRP-L10 Polyclonal Antibody detects endogenous levels encoded by nuclear genes and help in protein synthesis of MRP-L10 protein. within the mitochondrion. Mitochondrial ribosomes (mi- DATA: toribosomes) consist of a small 28S subunit and a large 39S subunit. They have an estimated 75% protein to rRNA composition compared to prokaryotic ribosomes, where this ratio is reversed. Another difference between mammalian mitoribosomes and prokaryotic ribosomes is that the latter contain a 5S rRNA. Among different spe- cies, the proteins comprising the mitoribosome differ greatly in sequence, and sometimes in biochemical prop- Western Blot analysis of various cells using MRP-L10 Polyclonal Anti- erties, which prevents easy recognition by sequence ho- body diluted at 1:1000 mology. This gene encodes a 39S subunit protein. Se- quence analysis identified three transcript variants that encode two different isoforms. A pseudogene corre- sponding to this gene is found on chromosome 5q. [pro- vided by RefSeq, Nov 2010], Product: Liquid in PBS containing 50% glycerol, 0.5% BSA and 0.02% sodium azide. Western blot analysis of 293T lysis using MRP-L10 antibody. Antibody Molecular Weight: was diluted at 1:1000 ~ 29 kDa Swiss-Prot: Q7Z7H8 Purification&Purity: The antibody was affinity-purified from rabbit antiserum by affinity-chromatography using epitope-specific im- munogen. Western blot analysis of lysates from 293, HepG2, and COLO cells, us- Applications: ing MRPL10 Antibody. The lane on the right is blocked with the syn- Western Blot: 1/500 - 1/2000. -

Enhanced Autophagic-Lysosomal Activity and Increased BAG3- Mediated Selective Macroautophagy As Adaptive Response of Neuronal Ce
bioRxiv preprint doi: https://doi.org/10.1101/580977; this version posted March 18, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Enhanced autophagic-lysosomal activity and increased BAG3- mediated selective macroautophagy as adaptive response of neuronal cells to chronic oxidative stress Debapriya Chakraborty1, Vanessa Felzen1, Christof Hiebel1, Elisabeth Stürner1, Natarajan Perumal2, Caroline Manicam2, Elisabeth Sehn3, Franz Grus2, Uwe Wolfrum3, Christian Behl1* 1Institute of Pathobiochemistry, University Medical Center Mainz of the Johannes Gutenberg University, 55099 Mainz, Germany 2Experimental and Translational Ophthalmology, University Medical Center Mainz, 55131 Mainz, Germany 3 Institute for Molecular Physiology, Johannes Gutenberg University, 55128 Mainz, Germany E-mail addresses of authors: D. Chakraborty: [email protected]; V. Felzen: [email protected]; C. Hiebel: [email protected]; E. Stürner: [email protected]; N. Perumal: nperumal@eye- research.org; C. Manicam: [email protected]; E. Sehn: sehn@uni- mainz.de; F. Grus: [email protected]; U. Wolfrum: [email protected]; *Correspondence to Christian Behl, PhD Institute of Pathobiochemistry University Medical Center of the Johannes Gutenberg University Duesbergweg 6, D-55099 Mainz [email protected] Tel.: +49-6131-39-25890; Fax: +49-6131-39-25792 1 bioRxiv preprint doi: https://doi.org/10.1101/580977; this version posted March 18, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Oxidative stress and a disturbed cellular protein homeostasis (proteostasis) belong to the most important hallmarks of aging and of neurodegenerative disorders. -
Mitochondrial Ribosomal Proteins: Candidate Genes for Mitochondrial Disease James E
March/April 2004 ⅐ Vol. 6 ⅐ No. 2 review Mitochondrial ribosomal proteins: Candidate genes for mitochondrial disease James E. Sylvester, PhD1, Nathan Fischel-Ghodsian, MD2, Edward B. Mougey, PhD1, and Thomas W. O’Brien, PhD3 Most of the energy requirement for cell growth, differentiation, and development is met by the mitochondria in the form of ATP produced by the process of oxidative phosphorylation. Human mitochondrial DNA encodes a total of 13 proteins, all of which are essential for oxidative phosphorylation. The mRNAs for these proteins are translated on mitochondrial ribosomes. Recently, the genes for human mitochondrial ribosomal proteins (MRPs) have been identified. In this review, we summarize their refined chromosomal location. It is well known that mutations in the mitochondrial translation system, i.e., ribosomal RNA and transfer RNA cause various pathologies. In this review, we suggest possible associations between clinical conditions and MRPs based on coincidence of genetic map data and chromosomal location. These MRPs may be candidate genes for the clinical condition or may act as modifiers of existing known gene mutations (mt-tRNA, mt-rRNA, etc.). Genet Med 2004:6(2):73–80. Key Words: mitochondrial, ribosomal proteins, oxidative phosphorylation, candidate genes, translation Most of the energy requirements for cell growth, differenti- THE MITORIBOSOME ation, and development are met by the mitochondrial ATP Human cells contain two genomes and two protein synthe- produced by the process of oxidative phosphorylation. Mito- sizing (translation) systems. The first is the nuclear genome of chondrial DNA encodes 13 essential proteins of this oxidative 3 ϫ 109 bp that has 30,000 to 40,000 genes coding a much phosphorylation system. -

Table 5. the List of 202 Derived Functional Modules
Table 5. The list of 202 derived functional modules MRP4 and MRPS5 are structural constituents of mitochondrial small ribosomal subunit involved in protein biosynthesis. Therefore, yet un- MRP4 annotated YDR036C (MRP5) is strongly suspected to have similar MRPS5 function. Their functional link can be checked by double or triple deletion experiments. YDR036C Parts of RNA polymerase II transcription mediator complex involved GAL11 in transcription from polymerase (Pol) II promoter. ROX3 SRB6 MED2 MED7 Parts of nuclear ubiquitin ligase complex involved in ubiquitin-protein ligase during G1/S and G2/M transitions of mitotic cell cycle. CDC53 SKP1 Parts of TRAPP complex involved in targeting and fusion of BET3 endoplasmic reticulum to Golgi vesicles. TRS20 CKA2 Alpha and beta subunits of casein kinase II complex involved in regulation of several cellular processes. CKB1 CKA1 CKB2 PRP45 and CEF1 are involved in pre-mRNA splicing with the PRP45 spliceosome. Therefore, yet unannotated YJU2 is strongly suspected to CEF1 have similar function. YJU2 GDI1 is a RAB GDP-dissociation inhibitor involved in vesicle- mediate transport, whereas YIF1 is part of COPII-coated vesicle YGL198W involved in ER to Golgi transport. Therefore, yet unannotated YGL161C YGL161C (YIP5) and YGL198W (YIP4) are strongly suspected to be involved in similar cellular functions. YGL161C and YGL198W are YIF1 not homologous to remaining two proteins. GDI1 Parts of chaperone ring complex located in the cytoplasm and assisting CCT5 in protein folding. CCT3 CCT2 TCP1 CCT6 Both mitogen-activated protein (MAP) kinase proteins involved in DIG1 down-regulation of invasive growth. It is known that each of them is viable, but double deletion results in loss of function.