Bartonella Infection in Sylvatic Small Mammals of Central Sweden
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genetic Diversity of Bartonella Species in Small Mammals in the Qaidam
www.nature.com/scientificreports OPEN Genetic diversity of Bartonella species in small mammals in the Qaidam Basin, western China Huaxiang Rao1, Shoujiang Li3, Liang Lu4, Rong Wang3, Xiuping Song4, Kai Sun5, Yan Shi3, Dongmei Li4* & Juan Yu2* Investigation of the prevalence and diversity of Bartonella infections in small mammals in the Qaidam Basin, western China, could provide a scientifc basis for the control and prevention of Bartonella infections in humans. Accordingly, in this study, small mammals were captured using snap traps in Wulan County and Ge’ermu City, Qaidam Basin, China. Spleen and brain tissues were collected and cultured to isolate Bartonella strains. The suspected positive colonies were detected with polymerase chain reaction amplifcation and sequencing of gltA, ftsZ, RNA polymerase beta subunit (rpoB) and ribC genes. Among 101 small mammals, 39 were positive for Bartonella, with the infection rate of 38.61%. The infection rate in diferent tissues (spleens and brains) (χ2 = 0.112, P = 0.738) and gender (χ2 = 1.927, P = 0.165) of small mammals did not have statistical diference, but that in diferent habitats had statistical diference (χ2 = 10.361, P = 0.016). Through genetic evolution analysis, 40 Bartonella strains were identifed (two diferent Bartonella species were detected in one small mammal), including B. grahamii (30), B. jaculi (3), B. krasnovii (3) and Candidatus B. gerbillinarum (4), which showed rodent-specifc characteristics. B. grahamii was the dominant epidemic strain (accounted for 75.0%). Furthermore, phylogenetic analysis showed that B. grahamii in the Qaidam Basin, might be close to the strains isolated from Japan and China. -

Abstract Pultorak, Elizabeth Lauren
ABSTRACT PULTORAK, ELIZABETH LAUREN. The Epidemiology of Lyme Disease and Bartonellosis in Humans and Animals. (Under the direction of Edward B. Breitschwerdt). The expansion of vector borne diseases in humans, a variety of mammalian hosts, and arthropod vectors draws attention to the need for enhanced diagnostic techniques for documenting infection in hosts, effective vector control, and treatment of individuals with associated diseases. Through improved diagnosis of vector-borne disease in both humans and animals, epidemiological studies to elucidate clinical associations or spatio-temporal relationships can be assessed. Veterinarians, through the use of the C6 peptide in the SNAP DX test kit, may be able to evaluate the changing epidemiology of borreliosis through their canine population. We developed a survey to evaluate the practices and perceptions of veterinarians in North Carolina regarding borreliosis in dogs across different geographic regions of the state. We found that veterinarians’ perception of the risk of borreliosis in North Carolina was consistent with recent scientific reports pertaining to geographic expansion of borreliosis in the state. Veterinarians should promote routine screening of dogs for Borrelia burgdorferi exposure as a simple, inexpensive form of surveillance in this transitional geographic region. We next conducted two separate studies to evaluate Bartonella spp. bacteremia or presence of antibodies against B. henselae, B. koehlerae, or B. vinsonii subsp. berkhoffii in 296 patients examined by a rheumatologist and 192 patients with animal exposure (100%) and recent animal bites and scratches (88.0%). Among 296 patients examined by a rheumatologist, prevalence of antibodies (185 [62%]) and Bartonella spp. bacteremia (122 [41.1%]) was high. -

Beating Chronic LYME
Section 1 Beating Chronic LYME Dr. Kevin Conners Fellowship in Integrative Cancer Therapy Fellowship in Anti-Aging, Regenerative, and Functional Medicine American Academy of Anti-Aging Medicine www.ConnersClinic.com From left to right: Larvae, Nymph, Female, Male Tick Tick in Nymph stage is the size of a poppy seed. Beating Chronic LYME Forward I used to live in the woods. My wife and four children at the time purchased 280 acres in Wisconsin and basically lived off the land. We grew most of our own food, were ‘off grid’ as we produced our own electricity through solar panels, and had to pump our water by hand. It was certainly a different way of life that prepared us for missionary work in Mexico. It was 1997 and at the time I had begun hearing about Lyme disease being a tick- born disorder. I had never seen a deer tick before moving to our ‘little house in the big woods’ but one thing was for certain – we had plenty of deer. They were as populous as the mosquitoes. To make a long story short, both my oldest daughter and I had contracted Lyme during our three year stay. I experienced the violent sickness of acute Lyme as well as a beautiful bulls-eye rash that made the diagnosis easy. I took just 3 days of antibiotics and since that was just the second time that I had ever taken a prescription medication in my life, they eradicated the disease effectively. My daughter on the other hand wasn’t so fortunate. She never had an acute illness and we never saw any rash therefore we didn’t catch the disease until it had advanced to Chronic Lyme Disease (CLD). -

Detection and Partial Molecular Characterization of Rickettsia and Bartonella from Southern African Bat Species
Detection and partial molecular characterization of Rickettsia and Bartonella from southern African bat species by Tjale Mabotse Augustine (29685690) Submitted in partial fulfillment of the requirements for the degree MAGISTER SCIENTIAE (MICROBIOLOGY) in the Department of Microbiology and Plant Pathology Faculty of Natural and Agricultural Sciences University of Pretoria Pretoria, South Africa Supervisor: Dr Wanda Markotter Co-supervisors: Prof Louis H. Nel Dr Jacqueline Weyer May, 2012 I declare that the thesis, which I hereby submit for the degree MSc (Microbiology) at the University of Pretoria, South Africa, is my own work and has not been submitted by me for a degree at another university ________________________________ Tjale Mabotse Augustine i Acknowledgements I would like send my sincere gratitude to the following people: Dr Wanda Markotter (University of Pretoria), Dr Jacqueline Weyer (National Institute for Communicable Diseases-National Health Laboratory Service) and Prof Louis H Nel (University of Pretoria) for their supervision and guidance during the project. Dr Jacqueline Weyer (Centre for Zoonotic and Emerging diseases (Previously Special Pathogens Unit), National Institute for Communicable Diseases (National Heath Laboratory Service), for providing the positive control DNA for Rickettsia and Dr Jenny Rossouw (Special Bacterial Pathogens Reference Unit, National Institute for Communicable Diseases-National Health Laboratory Service), for providing the positive control DNA for Bartonella. Dr Teresa Kearney (Ditsong Museum of Natural Science), Gauteng and Northern Region Bat Interest Group, Kwa-Zulu Natal Bat Interest Group, Prof Ara Monadjem (University of Swaziland), Werner Marias (University of Johannesburg), Dr Francois du Rand (University of Johannesburg) and Prof David Jacobs (University of Cape Town) for collection of blood samples. -

Bartonella Spp. Infection Rate and B. Grahamii in Ticks
LETTERS 8. Reif KE, Macaluso KR. Ecology which is common in Europe, may nlm.nih.gov/Blast.cgi) comparison to of Rickettsia felis: a review. J Med also transmit zoonotic Bartonella spp. published sequences. Entomol. 2009;46:723–36. http://dx.doi. org/10.1603/033.046.0402 Evidence of possible tick transmission On the basis of the amplicon- 9. Reif KE, Stout RW, Henry GC, Foil LD, of bartonellae to humans under natural specifi c melting temperature and DNA Macaluso KR. Prevalence and infection conditions was provided by Eskow et bands representing the specifi c size load dynamics of Rickettsia felis in al. (3) and Angelakis et al. (4), who of 249-bp after gel electrophoresis, actively feeding cat fl eas. PLoS ONE. 2008;3:e2805. http://dx.doi.org/10.1371/ identifi ed Bartonella spp. in tissue results of qPCR showed 100 (4.76%) journal.pone.0002805 samples of patients who were recently infected I. ricinus ticks (Table). 10. Mitchell CJ. The role of Aedes albopictus bitten by ticks. We determined the Positive results did not vary by as an arbovirus vector. Parassitologia. prevalence of Bartonella spp. in developmental tick stages; 4.84% 1995;37:109–13. questing I. ricinus ticks in the city (18/372) adult ticks (5.08% [9/177] Address for correspondence: Didier Raoult, of Hanover, Germany, which is female and 4.62% [9/195] male), URMITE, UMR CNRS 7278, IRD 198, nicknamed The Green Metropolis and 4.71% (80/1,698) nymphs, and 6.67% INSERM 1095, Faculté de Médecine, 27 Bd was selected the German Capital of (2/30) larvae were infected (Table). -

Bartonella Henselae
Maggi et al. Parasites & Vectors 2013, 6:101 http://www.parasitesandvectors.com/content/6/1/101 RESEARCH Open Access Bartonella henselae bacteremia in a mother and son potentially associated with tick exposure Ricardo G Maggi1,3*, Marna Ericson2, Patricia E Mascarelli1, Julie M Bradley1 and Edward B Breitschwerdt1 Abstract Background: Bartonella henselae is a zoonotic, alpha Proteobacterium, historically associated with cat scratch disease (CSD), but more recently associated with persistent bacteremia, fever of unknown origin, arthritic and neurological disorders, and bacillary angiomatosis, and peliosis hepatis in immunocompromised patients. A family from the Netherlands contacted our laboratory requesting to be included in a research study (NCSU-IRB#1960), designed to characterize Bartonella spp. bacteremia in people with extensive arthropod or animal exposure. All four family members had been exposed to tick bites in Zeeland, southwestern Netherlands. The mother and son were exhibiting symptoms including fatigue, headaches, memory loss, disorientation, peripheral neuropathic pain, striae (son only), and loss of coordination, whereas the father and daughter were healthy. Methods: Each family member was tested for serological evidence of Bartonella exposure using B. vinsonii subsp. berkhoffii genotypes I-III, B. henselae and B. koehlerae indirect fluorescent antibody assays and for bacteremia using the BAPGM enrichment blood culture platform. Results: The mother was seroreactive to multiple Bartonella spp. antigens and bacteremia was confirmed by PCR amplification of B. henselae DNA from blood, and from a BAPGM blood agar plate subculture isolate. The son was not seroreactive to any Bartonella sp. antigen, but B. henselae DNA was amplified from several blood and serum samples, from BAPGM enrichment blood culture, and from a cutaneous striae biopsy. -

Characterizing the Molecular Biology of a Bacteriophage-Like Particle from Bartonella Bacilliformis
University of Montana ScholarWorks at University of Montana Graduate Student Theses, Dissertations, & Professional Papers Graduate School 1999 Characterizing the molecular biology of a bacteriophage-like particle from Bartonella bacilliformis Kent D. Barbian The University of Montana Follow this and additional works at: https://scholarworks.umt.edu/etd Let us know how access to this document benefits ou.y Recommended Citation Barbian, Kent D., "Characterizing the molecular biology of a bacteriophage-like particle from Bartonella bacilliformis" (1999). Graduate Student Theses, Dissertations, & Professional Papers. 6634. https://scholarworks.umt.edu/etd/6634 This Thesis is brought to you for free and open access by the Graduate School at ScholarWorks at University of Montana. It has been accepted for inclusion in Graduate Student Theses, Dissertations, & Professional Papers by an authorized administrator of ScholarWorks at University of Montana. For more information, please contact [email protected]. i I S Maureen and Mike MANSFIELD LIBRARY The University of IVIONTANA Permission is granted by the author to reproduce this material in its entirety, provided that this material is used for scholarly purposes and is properly cited in published works and reports. ** Please check ’’Yes'* or ”No” and provide signature ** Yes, I grant permission X" No, I do not grant permission _____ Author's Signature Date Any copying for conunercial purposes or financial gain may be undertaken only with the author's explicit consent. Characterizing the Molecular Biology of a Bacteriophage-Like Particle FromBartonella bacilliformis by Kent D. Barbian B.A,, The University of Montana, 1997 Presented in partial fulfillment of the requirements for the degree of Master of Science in Microbiology The University of Montana 1999 Approved by: Committee Chair Dean, Graduate School Date UMI Number: EP37435 All rights reserved INFORMATION TO ALL USERS The quality of this reproduction is dependent upon the quality of the copy submitted. -
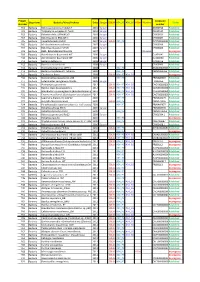
Project Number Organisms Bacteria/Virus/Archaea Date
Project_ Accession Organisms Bacteria/Virus/Archaea Date Sanger SOLiD 454_PE 454_SG PGM Illumina Status Number number P01 Bacteria Rickettsia conorii str.Malish 7 2001 Sanger AE006914 Published P02 Bacteria Tropheryma whipplei str.Twist 2003 Sanger AE014184 Published P03 Bacteria Rickettsia felis URRWXCal2 2005 Sanger CP000053 Published P04 Bacteria Rickettsia bellii RML369-C 2006 Sanger CP000087 Published P05 Bacteria Coxiella burnetii CB109 2007 Sanger SOLiD 454_PE AKYP00000000 Published P06 Bacteria Minibacterium massiliensis 2007 Sanger CP000269 Published P07 Bacteria Rickettsia massiliae MTU5 2007 Sanger CP000683 Published P08 Bacteria BaBL=Bête à Bernard Lascola 2007 Illumina In progress P09 Bacteria Acinetobacter baumannii AYE 2006 Sanger CU459141 Published P10 Bacteria Acinetobacter baumannii SDF 2006 Sanger CU468230 Published P11 Bacteria Borrelia duttonii Ly 2008 Sanger CP000976 Published P12 Bacteria Borrelia recurrentis A1 2008 Sanger CP000993 Published P13 Bacteria Francisella tularensis URFT1 2008 454_PE ABAZ00000000Published P14 Bacteria Borrelia crocidurae str. Achema 2009 454_PE PRJNA162335 Published P15 Bacteria Citrobacter koseri 2009 SOLiD 454_PE 454_SG In progress P16 Bacteria Diplorickettsia massiliensis 20B 2009 454_PE PRJNA86907 Published P17 Bacteria Enterobacter aerogenes EA1509E 2009 Sanger FO203355 Published P18 Bacteria Actinomyces grossensis 2012 SOLiD 454_PE 454_SG CAGY00000000Published P19 Bacteria Bacillus massiliosenegalensis 2012 SOLiD 454_PE 454_SG CAHJ00000000 Published P20 Bacteria Brevibacterium senegalensis -

Human Bartonellosis: an Underappreciated Public Health Problem?
Tropical Medicine and Infectious Disease Review Human Bartonellosis: An Underappreciated Public Health Problem? Mercedes A. Cheslock and Monica E. Embers * Division of Immunology, Tulane National Primate Research Center, Tulane University Health Sciences, Covington, LA 70433, USA; [email protected] * Correspondence: [email protected]; Tel.: +(985)-871-6607 Received: 24 March 2019; Accepted: 16 April 2019; Published: 19 April 2019 Abstract: Bartonella spp. bacteria can be found around the globe and are the causative agents of multiple human diseases. The most well-known infection is called cat-scratch disease, which causes mild lymphadenopathy and fever. As our knowledge of these bacteria grows, new presentations of the disease have been recognized, with serious manifestations. Not only has more severe disease been associated with these bacteria but also Bartonella species have been discovered in a wide range of mammals, and the pathogens’ DNA can be found in multiple vectors. This review will focus on some common mammalian reservoirs as well as the suspected vectors in relation to the disease transmission and prevalence. Understanding the complex interactions between these bacteria, their vectors, and their reservoirs, as well as the breadth of infection by Bartonella around the world will help to assess the impact of Bartonellosis on public health. Keywords: Bartonella; vector; bartonellosis; ticks; fleas; domestic animals; human 1. Introduction Several Bartonella spp. have been linked to emerging and reemerging human diseases (Table1)[ 1–5]. These fastidious, gram-negative bacteria cause the clinically complex disease known as Bartonellosis. Historically, the most common causative agents for human disease have been Bartonella bacilliformis, Bartonella quintana, and Bartonella henselae. -

Ru 2015 150 263 a (51) Мпк A61k 31/155 (2006.01)
РОССИЙСКАЯ ФЕДЕРАЦИЯ (19) (11) (13) RU 2015 150 263 A (51) МПК A61K 31/155 (2006.01) ФЕДЕРАЛЬНАЯ СЛУЖБА ПО ИНТЕЛЛЕКТУАЛЬНОЙ СОБСТВЕННОСТИ (12) ЗАЯВКА НА ИЗОБРЕТЕНИЕ (21)(22) Заявка: 2015150263, 01.05.2014 (71) Заявитель(и): НЕОКУЛИ ПТИ ЛТД (AU) Приоритет(ы): (30) Конвенционный приоритет: (72) Автор(ы): 01.05.2013 AU 2013901517 ПЕЙДЖ Стефен (AU), ГАРГ Санджай (AU) (43) Дата публикации заявки: 06.06.2017 Бюл. № 16 RU (85) Дата начала рассмотрения заявки PCT на национальной фазе: 01.12.2015 (86) Заявка PCT: AU 2014/000480 (01.05.2014) 2015150263 (87) Публикация заявки PCT: WO 2014/176634 (06.11.2014) Адрес для переписки: 190000, Санкт-Петербург, Box-1125, "ПАТЕНТИКА" A (54) СПОСОБЫ ЛЕЧЕНИЯ БАКТЕРИАЛЬНЫХ ИНФЕКЦИЙ (57) Формула изобретения 1. Способ лечения или профилактики бактериальной колонизации или инфекции у субъекта, включающий стадию: введения субъекту терапевтически эффективного количества робенидина или его терапевтически приемлемой соли, причем указанная A бактериальная колонизация или инфекция вызвана бактериальным агентом. 2. Способ по п. 1, отличающийся тем, что субъект выбран из группы, включающей: человека, животных, принадлежащих видам семейства псовых, кошачьих, крупного рогатого скота, овец, коз, свиней, птиц, рыб и лошадей. 3. Способ по п. 1, отличающийся тем, что робенидин вводят субъекту в дозе в диапазоне от 0,1 до 250 мг/кг массы тела. 4. Способ по любому из пп. 1-3, отличающийся тем, что бактериальный агент является 2015150263 грамположительным. 5. Способ по п. 4, отличающийся тем, что бактериальный агент выбран из -

Biosafety Guidelines for Contained Use of Genetically Modified Microorganisms at Pilot and Industrial Scales
Biosafety Guidelines for Contained Use of Genetically Modified Microorganisms at Pilot and Industrial Scales TECHNICAL BIOSAFETY COMMITTEE (TBC) NATIONAL CENTER FOR GENETIC ENGINEERING AND BIOTECHNOLOGY (BIOTEC) NATIONAL SCIENCE AND TECHNOLOGY DEVELOPMENT AGENCY (NSTDA) MINISTRY OF SCIENCE AND TECHNOLOGY (MOST) 2015 Biosafety Guidelines for Contained Use of Genetically Modified Microorganisms at Pilot and Industrial Scales TECHNICAL BIOSAFETY COMMITTEE (TBC) NATIONAL CENTER FOR GENETIC ENGINEERING AND BIOTECHNOLOGY (BIOTEC) NATIONAL SCIENCE AND TECHNOLOGY DEVELOPMENT AGENCY (NSTDA) MINISTRY OF SCIENCE AND TECHNOLOGY (MOST) 2015 Biosafety Guidelines for Contained Use of Genetically Modified Microorganisms at Pilot and Industrial Scales Technical Biosafety Committee National Center for Genetic Engineering and Biotechnology National Science and Technology Development Agency (NSTDA) © National Center for Genetic Engineering and Biotechnology 2015 ISBN : 978-616-12-0386-3 Tel : +66(0)2-564-6700 Fax : +66(0)2-564-6703 E-mail : [email protected] URL : http://www.biotec.or.th Printing House : P.A. Living Printing Co.,Ltd 4 Soi Sirintron 7 Road Sirintron District Bangplad Province Bangkok 10700 Tel : +66(0)2-881 9890 Fax : +66(0)2-881 9894 Preface Genetically Modified Microorganisms (GMMs) were first used in B.E. 2525 to produce insulin in industrial medicine. Currently, GMMs are used in various industries, such as the food, pharmaceutical and bioplastic industries, to manufacture a number of important consumer products. To ensure operator and environmental safety, the Technical Biosafety Committee (TBC) of the National Center for Genetic Engineering and Biotechnology (BIOTEC), the National Science and Technology Development Agency (NSTDA), has prepared guidelines for GMM work, publishing “Biosafety Guidelines for Contained Use of Genetically Modified Microorganisms at Pilot and Industrial Scales” in B.E. -

Evolution of Methanotrophy in the Beijerinckiaceae&Mdash
The ISME Journal (2014) 8, 369–382 & 2014 International Society for Microbial Ecology All rights reserved 1751-7362/14 www.nature.com/ismej ORIGINAL ARTICLE The (d)evolution of methanotrophy in the Beijerinckiaceae—a comparative genomics analysis Ivica Tamas1, Angela V Smirnova1, Zhiguo He1,2 and Peter F Dunfield1 1Department of Biological Sciences, University of Calgary, Calgary, Alberta, Canada and 2Department of Bioengineering, School of Minerals Processing and Bioengineering, Central South University, Changsha, Hunan, China The alphaproteobacterial family Beijerinckiaceae contains generalists that grow on a wide range of substrates, and specialists that grow only on methane and methanol. We investigated the evolution of this family by comparing the genomes of the generalist organotroph Beijerinckia indica, the facultative methanotroph Methylocella silvestris and the obligate methanotroph Methylocapsa acidiphila. Highly resolved phylogenetic construction based on universally conserved genes demonstrated that the Beijerinckiaceae forms a monophyletic cluster with the Methylocystaceae, the only other family of alphaproteobacterial methanotrophs. Phylogenetic analyses also demonstrated a vertical inheritance pattern of methanotrophy and methylotrophy genes within these families. Conversely, many lateral gene transfer (LGT) events were detected for genes encoding carbohydrate transport and metabolism, energy production and conversion, and transcriptional regulation in the genome of B. indica, suggesting that it has recently acquired these genes. A key difference between the generalist B. indica and its specialist methanotrophic relatives was an abundance of transporter elements, particularly periplasmic-binding proteins and major facilitator transporters. The most parsimonious scenario for the evolution of methanotrophy in the Alphaproteobacteria is that it occurred only once, when a methylotroph acquired methane monooxygenases (MMOs) via LGT.