An Emerging Pathogen from Rotted Chestnut in China: Gnomoniopsis Daii Sp
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

CHESTNUT (CASTANEA Spp.) CULTIVAR EVALUATION for COMMERCIAL CHESTNUT PRODUCTION
CHESTNUT (CASTANEA spp.) CULTIVAR EVALUATION FOR COMMERCIAL CHESTNUT PRODUCTION IN HAMILTON COUNTY, TENNESSEE By Ana Maria Metaxas Approved: James Hill Craddock Jennifer Boyd Professor of Biological Sciences Assistant Professor of Biological and Environmental Sciences (Director of Thesis) (Committee Member) Gregory Reighard Jeffery Elwell Professor of Horticulture Dean, College of Arts and Sciences (Committee Member) A. Jerald Ainsworth Dean of the Graduate School CHESTNUT (CASTANEA spp.) CULTIVAR EVALUATION FOR COMMERCIAL CHESTNUT PRODUCTION IN HAMILTON COUNTY, TENNESSEE by Ana Maria Metaxas A Thesis Submitted to the Faculty of the University of Tennessee at Chattanooga in Partial Fulfillment of the Requirements for the Degree of Master of Science in Environmental Science May 2013 ii ABSTRACT Chestnut cultivars were evaluated for their commercial applicability under the environmental conditions in Hamilton County, TN at 35°13ꞌ 45ꞌꞌ N 85° 00ꞌ 03.97ꞌꞌ W elevation 230 meters. In 2003 and 2004, 534 trees were planted, representing 64 different cultivars, varieties, and species. Twenty trees from each of 20 different cultivars were planted as five-tree plots in a randomized complete block design in four blocks of 100 trees each, amounting to 400 trees. The remaining 44 chestnut cultivars, varieties, and species served as a germplasm collection. These were planted in guard rows surrounding the four blocks in completely randomized, single-tree plots. In the analysis, we investigated our collection predominantly with the aim to: 1) discover the degree of acclimation of grower- recommended cultivars to southeastern Tennessee climatic conditions and 2) ascertain the cultivars’ ability to survive in the area with Cryphonectria parasitica and other chestnut diseases and pests present. -

Frank Meyer, Isabel Shipley Cunningham Agricultural Explorer
Frank Meyer, Isabel Shipley Cunningham Agricultural Explorer For 60 years the work of Frank N. Meyer has 2,500 pages of his letters tell of his journeys remained a neglected segment of America’s and the plants he collected, and the USDA heritage. Now, as people are becoming con- Inventory of Seeds and Plants Imported con- cerned about feeding the world’s growing tains descriptions of his introductions. population and about the loss of genetic di- Until recently little was known about the versity of crops, Meyer’s accomplishments first 25 years of Meyer’s life, when he lived have a special relevance. Entering China in in Amsterdam and was called Frans Meijer. 1905, near the dawn of the single era when Dutch sources reveal that he was bom into a explorers could travel freely there, he be- loving family in 1875. Frans was a quiet boy, came the first plant hunter to represent a who enjoyed taking long walks, reading government and to search primarily for eco- about distant lands, and working in his fami- nomically useful plants rather than orna- ly’s small garden. By the time he had mentals. No one before him had spent 10 fimshed elementary school, he knew that ’ years crossing the mountains, deserts, farms, wanted to be a world traveler who studied he ’,if and forests of Asia in search of fruits, nuts, plants; however, his parents could not afford vegetables, grains, and fodder crops; no one to give him further education. When he was has done so since. 14 years old, he found work as a gardener’s During four plant-hunting expeditions to helper at the Amsterdam Botanical Garden. -

Castanea Mollissima Blume): the Roots of Nut Tree Domestication
ORIGINAL RESEARCH published: 25 June 2018 doi: 10.3389/fpls.2018.00810 Signatures of Selection in the Genomes of Chinese Chestnut (Castanea mollissima Blume): The Roots of Nut Tree Domestication Nicholas R. LaBonte 1*, Peng Zhao 2 and Keith Woeste 3 1 Department of Crop Sciences, University of Illinois Urbana-Champaign, Urbana, IL, United States, 2 Key Laboratory of Resource Biology and Biotechnology in Western China, Ministry of Education, College of Life Sciences, Northwest University, Xi’an, China, 3 Hardwood Tree Improvement and Regeneration Center, Northern Research Station, USDA Forest Service, West Lafayette, IN, United States Chestnuts (Castanea) are major nut crops in East Asia and southern Europe, and are unique among temperate nut crops in that the harvested seeds are starchy rather than oily. Chestnut species have been cultivated for three millennia or more in China, so it is likely that artificial selection has affected the genome of orchard-grown chestnuts. The genetics of Chinese chestnut (Castanea mollissima Blume) domestication are also of Edited by: interest to breeders of hybrid American chestnut, especially if the low-growing, branching S. Hong Lee, habit of Chinese chestnut, an impediment to American chestnut restoration, is partly University of South Australia, Australia the result of artificial selection. We resequenced genomes of wild and orchard-derived Reviewed by: Guo-Bo Chen, Chinese chestnuts and identified selective sweeps based on pooled whole-genome SNP Zhejiang Provincial People’s Hospital, datasets. We present candidate gene loci for chestnut domestication and discuss the China potential phenotypic effects of candidate loci, some of which may be useful genes for Chaeyoung Lee, Soongsil University, South Korea chestnut improvement in Asia and North America. -

Chestnut Growers' Guide to Site Selection and Environmental Stress
This idyllic orchard has benefited from good soil and irrigation. Photo by Tom Saielli Chestnut Growers’ Guide to Site Selection and Environmental Stress By Elsa Youngsteadt American chestnuts are tough, efficient trees that can reward their growers with several feet of growth per year. They’ll survive and even thrive under a range of conditions, but there are a few deal breakers that guarantee sickly, slow-growing trees. This guide, intended for backyard and small-orchard growers, will help you avoid these fatal mistakes and choose planting sites that will support strong, healthy trees. You’ll know you’ve done well when your chestnuts are still thriving a few years after planting. By then, they’ll be strong enough to withstand many stresses, from drought to a caterpillar outbreak, with much less human help. Soil Soil type is the absolute, number-one consideration when deciding where—or whether—to plant American chestnuts. These trees demand well-drained, acidic soil with a sandy to loamy texture. Permanently wet, basic, or clay soils are out of the question. So spend some time getting to know your dirt before launching a chestnut project. Dig it up, roll it between your fingers, and send in a sample for a soil test. Free tests are available through most state extension programs, and anyone can send a sample to the Penn State Agricultural Analytical Services Lab (which TACF uses) for a small fee. More information can be found at http://agsci.psu.edu/aasl/soil-testing. There are several key factors to look for. The two-foot-long taproot on this four- Acidity year-old root system could not have The ideal pH for American chestnut is 5.5, with an acceptable range developed in shallow soils, suggesting from about 4.5 to 6.5. -
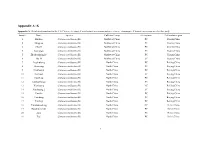
Appendix A~K
Appendix A~K Appendix A. Detailed information for the 146 Chinese chestnut (C.mollissima) accessions and nine chinese chinquapin (C.henryi) accessions used in this study Sample Name Species Cultivars Group Germplasm Cultivation region 1 Huijian Castanea mollissima Bl. Northwest China PC Shanxi,China 2 Mingjian Castanea mollissima Bl. Northwest China PC Shanxi,China 3 Chunli Castanea mollissima Bl. Northwest China PC Shanxi,China 4 Zuohongli Castanea mollissima Bl. Northwest China PC Shanxi,China 5 Zhenbashuangjie Castanea mollissima Bl. Northwest China PC Shanxi,China 6 zha 18 Castanea mollissima Bl. Northwest China LC Shanxi,China 7 Jingshuhong Castanea mollissima Bl. North China PC Beijing,China 8 Huaixiang Castanea mollissima Bl. North China PC Beijing,China 9 Huaihuang Castanea mollissima Bl. North China PC Beijing,China 10 Huzhaoli Castanea mollissima Bl. North China LC Beijing,China 11 Huaifeng Castanea mollissima Bl. North China PC Beijing,China 12 Yanshanhongli Castanea mollissima Bl. North China PC Beijing,China 13 Xiazhuang Castanea mollissima Bl. North China LC Beijing,China 14 Xinzhuang 2 Castanea mollissima Bl. North China LC Beijing,China 15 Huaijiu Castanea mollissima Bl. North China PC Beijing,China 16 Yanchang Castanea mollissima Bl. North China PC Beijing,China 17 Yanfeng Castanea mollissima Bl. North China PC Beijing,China 18 Yanshanzaofeng Castanea mollissima Bl. North China NC Hebei,China 19 Donglimingzhu Castanea mollissima Bl. North China PC Hebei,China 20 Zipo Castanea mollissima Bl. North China PC Hebei,China 21 Tasi Castanea mollissima Bl. North China PC Hebei,China 1 22 Zundali Castanea mollissima Bl. North China PC Hebei,China 23 Duanzhiyabian Castanea mollissima Bl. -

Castanea Mollissima
Castanea mollissima As the American chestnut struggles with disease, the blight- resistant Chinese chestnut is quickly gaining popularity. The sweet-tasting nuts are often roasted for holiday eating and have been made famous in turkey stuffing recipes across the country. But this is more than a nut tree. The shade of its spreading canopy is dense, providing relief in the hot, dry climates the Chinese chestnut does well in. Hardiness Zones: The chinese chestnut can be expected to grow in Hardiness Zones 4-8. Tree Type: This is a nut-producing tree, yielding nuts for human and wildlife consumption. Mature Size: The Chinese chestnut grows to a height of 40-60' and a spread of 40-60' at maturity. Growth Rate: This tree grows at a slow to medium rate, with height increases of anywhere from less than 12" to 24" per year. Sun Preference: Full sun is the ideal condition for this tree, meaning it should get at least six hours of direct, unfiltered sunlight each day. Soil Preference: The Chinese chestnut grows in acidic, loamy, moist, sandy, well-drained, and clay soils. It is drought-tolerant. Attributes This tree: Should be planted in pairs or groups to ensure pollination. 1 Yields a ripened nut crop mid/late September through October. A prickly 2-3 /2" seed husk en- closes 1-4 nuts. The nuts are large, meaty, crisp, and sweet, although less sweet than American chestnuts. Begins to bear nuts in 4-5 years if grown from seed. Provides dense shade with a handsome, spreading canopy. Has wood that is very durable and resistant to rot. -

Alien Invasive Species and International Trade
Forest Research Institute Alien Invasive Species and International Trade Edited by Hugh Evans and Tomasz Oszako Warsaw 2007 Reviewers: Steve Woodward (University of Aberdeen, School of Biological Sciences, Scotland, UK) François Lefort (University of Applied Science in Lullier, Switzerland) © Copyright by Forest Research Institute, Warsaw 2007 ISBN 978-83-87647-64-3 Description of photographs on the covers: Alder decline in Poland – T. Oszako, Forest Research Institute, Poland ALB Brighton – Forest Research, UK; Anoplophora exit hole (example of wood packaging pathway) – R. Burgess, Forestry Commission, UK Cameraria adult Brussels – P. Roose, Belgium; Cameraria damage medium view – Forest Research, UK; other photographs description inside articles – see Belbahri et al. Language Editor: James Richards Layout: Gra¿yna Szujecka Print: Sowa–Print on Demand www.sowadruk.pl, phone: +48 022 431 81 40 Instytut Badawczy Leœnictwa 05-090 Raszyn, ul. Braci Leœnej 3, phone [+48 22] 715 06 16 e-mail: [email protected] CONTENTS Introduction .......................................6 Part I – EXTENDED ABSTRACTS Thomas Jung, Marla Downing, Markus Blaschke, Thomas Vernon Phytophthora root and collar rot of alders caused by the invasive Phytophthora alni: actual distribution, pathways, and modeled potential distribution in Bavaria ......................10 Tomasz Oszako, Leszek B. Orlikowski, Aleksandra Trzewik, Teresa Orlikowska Studies on the occurrence of Phytophthora ramorum in nurseries, forest stands and garden centers ..........................19 Lassaad Belbahri, Eduardo Moralejo, Gautier Calmin, François Lefort, Jose A. Garcia, Enrique Descals Reports of Phytophthora hedraiandra on Viburnum tinus and Rhododendron catawbiense in Spain ..................26 Leszek B. Orlikowski, Tomasz Oszako The influence of nursery-cultivated plants, as well as cereals, legumes and crucifers, on selected species of Phytophthopra ............30 Lassaad Belbahri, Gautier Calmin, Tomasz Oszako, Eduardo Moralejo, Jose A. -

RAL COLOR CHART ***** This Chart Is to Be Used As a Guide Only. Colors May Appear Slightly Different ***** Green Beige Purple V
RAL COLOR CHART ***** This Chart is to be used as a guide only. Colors May Appear Slightly Different ***** RAL 1000 Green Beige RAL 4007 Purple Violet RAL 7008 Khaki Grey RAL 4008 RAL 7009 RAL 1001 Beige Signal Violet Green Grey Tarpaulin RAL 1002 Sand Yellow RAL 4009 Pastel Violet RAL 7010 Grey RAL 1003 Signal Yellow RAL 5000 Violet Blue RAL 7011 Iron Grey RAL 1004 Golden Yellow RAL 5001 Green Blue RAL 7012 Basalt Grey Ultramarine RAL 1005 Honey Yellow RAL 5002 RAL 7013 Brown Grey Blue RAL 1006 Maize Yellow RAL 5003 Saphire Blue RAL 7015 Slate Grey Anthracite RAL 1007 Chrome Yellow RAL 5004 Black Blue RAL 7016 Grey RAL 1011 Brown Beige RAL 5005 Signal Blue RAL 7021 Black Grey RAL 1012 Lemon Yellow RAL 5007 Brillant Blue RAL 7022 Umbra Grey Concrete RAL 1013 Oyster White RAL 5008 Grey Blue RAL 7023 Grey Graphite RAL 1014 Ivory RAL 5009 Azure Blue RAL 7024 Grey Granite RAL 1015 Light Ivory RAL 5010 Gentian Blue RAL 7026 Grey RAL 1016 Sulfer Yellow RAL 5011 Steel Blue RAL 7030 Stone Grey RAL 1017 Saffron Yellow RAL 5012 Light Blue RAL 7031 Blue Grey RAL 1018 Zinc Yellow RAL 5013 Cobolt Blue RAL 7032 Pebble Grey Cement RAL 1019 Grey Beige RAL 5014 Pigieon Blue RAL 7033 Grey RAL 1020 Olive Yellow RAL 5015 Sky Blue RAL 7034 Yellow Grey RAL 1021 Rape Yellow RAL 5017 Traffic Blue RAL 7035 Light Grey Platinum RAL 1023 Traffic Yellow RAL 5018 Turquiose Blue RAL 7036 Grey RAL 1024 Ochre Yellow RAL 5019 Capri Blue RAL 7037 Dusty Grey RAL 1027 Curry RAL 5020 Ocean Blue RAL 7038 Agate Grey RAL 1028 Melon Yellow RAL 5021 Water Blue RAL 7039 Quartz Grey -

Diseases of Trees in the Great Plains
United States Department of Agriculture Diseases of Trees in the Great Plains Forest Rocky Mountain General Technical Service Research Station Report RMRS-GTR-335 November 2016 Bergdahl, Aaron D.; Hill, Alison, tech. coords. 2016. Diseases of trees in the Great Plains. Gen. Tech. Rep. RMRS-GTR-335. Fort Collins, CO: U.S. Department of Agriculture, Forest Service, Rocky Mountain Research Station. 229 p. Abstract Hosts, distribution, symptoms and signs, disease cycle, and management strategies are described for 84 hardwood and 32 conifer diseases in 56 chapters. Color illustrations are provided to aid in accurate diagnosis. A glossary of technical terms and indexes to hosts and pathogens also are included. Keywords: Tree diseases, forest pathology, Great Plains, forest and tree health, windbreaks. Cover photos by: James A. Walla (top left), Laurie J. Stepanek (top right), David Leatherman (middle left), Aaron D. Bergdahl (middle right), James T. Blodgett (bottom left) and Laurie J. Stepanek (bottom right). To learn more about RMRS publications or search our online titles: www.fs.fed.us/rm/publications www.treesearch.fs.fed.us/ Background This technical report provides a guide to assist arborists, landowners, woody plant pest management specialists, foresters, and plant pathologists in the diagnosis and control of tree diseases encountered in the Great Plains. It contains 56 chapters on tree diseases prepared by 27 authors, and emphasizes disease situations as observed in the 10 states of the Great Plains: Colorado, Kansas, Montana, Nebraska, New Mexico, North Dakota, Oklahoma, South Dakota, Texas, and Wyoming. The need for an updated tree disease guide for the Great Plains has been recog- nized for some time and an account of the history of this publication is provided here. -

Molecular Characterization of Strawberry Pathogen Gnomonia Fragariae and Its Genetic Relatedness to Other Gnomonia Species and Members of Diaporthales
mycological research 111 (2007) 603–614 available at www.sciencedirect.com journal homepage: www.elsevier.com/locate/mycres Molecular characterization of strawberry pathogen Gnomonia fragariae and its genetic relatedness to other Gnomonia species and members of Diaporthales Inga MOROCˇKOa,b,*, Jamshid FATEHIa aMASE Laboratories AB, Box 148, S-751 04, Uppsala, Sweden bDepartment of Forest Mycology and Pathology, Swedish University of Agricultural Sciences, Box 7026, S-750 07, Uppsala, Sweden article info abstract Article history: Gnomonia fragariae is a poorly studied ascomycete belonging to Diaporthales. Originally Received 11 September 2006 G. fragariae was considered a saprophyte occurring on dead tissues of strawberry plants. Received in revised form Recently this fungus was found in Latvia and Sweden, and it was proven to be the cause 14 February 2007 of severe root rot and petiole blight of strawberry. Thirteen isolates of this pathogen and Accepted 9 March 2007 several other Gnomonia species occurring on rosaceous hosts were characterized by molec- Published online 19 March 2007 ular analysis using nucleotide sequences of partial LSU rRNA gene and the total ITS region. Corresponding Editor: The homologous regions from relevant diaporthalean taxa available in the GenBank were David L. Hawksworth also included and compared with the taxa sequenced in this study. Phylogenetic analyses revealed that G. fragariae, G. rubi, and Gnomonia sp. (CBS 850.79) were genetically different Keywords: from G. gnomon, the type species of the genus, and other members of Gnomoniaceae. The Fragaria analyses showed that G. fragariae and Hapalocystis were genetically very closely related, Gnomoniaceae forming a phylogenetic clade, which is possibly presenting a new family in the Diaporthales. -

Download Colour Chart
DIRECT DYE * AMMONIA FREE * PEROXIDE FREE DIRECTIONS: CUSTOMISED COLOUR MAINTENANCE APPLICATION MIXING TIMING refer to menu for depending on hair condition suggested formulas and desired colour intensity colour maintenance add 80g fab pro 3 minutes conditioner formula (direct dye match and maintain colour or direct dye and in-between salon visits. conditioner) to 200ml conditioner base. shake well. MATCH IT * MIX IT * TAKE IT AWAY DIRECTIONS: COLOUR SERVICE 1. identify the level, tone and length of hair. 2. select the appropriate colour formula from the mixologist menu, ensuring the formula works with the lightest level of hair. 3. measure and mix your formula. 4. prepare hair by shampooing, towel-dry hair evenly, then detangle and comb through. APPLICATION MIXING TIMING refer to menu for depending on hair condition suggested formulas and desired colour intensity colour refresh mix fab pro direct 5 – 15 minutes refresh existing hair colour dyes together. on mid-lengths and ends in-between all colour services (permanent, demi and semi- permanent colour). colour fill* mix fab pro direct 5 – 15 minutes darken lighter hair quickly, dyes together. without damage. *for filling formulas, see fab pro fill chart or visit evohair.com. colour tone* / pastels mix fab pro direct 5 – 15 minutes ideal for colour toning and dyes together or with can be diluted to create pastel shades. conditioner base. *for extremely porous hair, you may need to dilute fab pro direct dye using conditioner base. DIRECT DYES LEVEL 5 LEVEL 7 LEVEL 10 colour results are determined by the level, tone and formula you use. in order to accurately predict a colour result, you need to understand how the existing level and tone will contribute to the result; the lighter the level, the more intense the result. -

Color Chart.Pdf
® Finishing Products Division of RPM Wood Finishes Group Inc. Color Chart The Original Touch Up Company™ Made in the USA Color Chart ® Finishing Products Division of RPM Wood Finishes Group, Inc. Index Aerosols 1-5 Ultra® Classic Toner & Tone Finish Toner 1-3 Colored Lacquer Enamel 3-5 Shadow Toner 5 Touch-Up Markers/Pencils 5-15 Ultra® Mark Markers 5-9 3 in 1 Repair Stick 9 Pro-Mark® Markers 9-10 Quik-Tip™ Markers 10-11 Background Marker Touch-Up & Background Marker Glaze Hang-Up 11-13 Artisan Glaze Markers 13 Vinyl Marker Glaze Hang-Up 14 Brush Tip Graining Markers 14 Accent Pencils 15 Blend-Its 15 Fillers 15-29 Quick Fill® Burn-In Sticks 15-16 Edging/Low Heat Sticks 16 E-Z Flow™ Burn-In Sticks 16-17 PlaneStick® Burn-In Sticks 17-18 Fil-Stik® Putty Sticks 18-25 Hard Fill & Hard Fill Plus 25-27 PermaFill™ 27 Epoxy Putty Sticks 27-28 Patchal® Puttys 28-29 Knot Filler 29 Fil-O-Wood™ Wood Putty Tubes 29 Color Replacement 30-31 Blendal® Sticks 30 Sand Thru Sticks 30-31 Blendal® Powder Stains 31 Bronzing Powders 31 Dye Stains 32 Ultra® Penetrating & Architectural Ultra® Penetrating Stain 32 Dye Concentrate 32 Pigmented Stains 32-34 Wiping Wood™, Architectural Wiping Stain & Wiping Wood™ Stain Aerosols 32-33 Designer Series Stain, Designer Series Radiant Stain 33-34 Glazes 34 Finisher’s Glaze™ Glazing Stain & Aerosols 34 Break-A-Way™ Glaze & Aerosols 34 Leather Repair 35-37 E-Z Flow™ Leather Markers 35 Leather/Vinyl Markers 35 Leather/Vinyl Fil Sticks 35-36 Leather Repair Basecoat Aerosols 36 Leather Repair Toner Aerosols 36 Leather Repair Color Adjuster Aerosols 37 Touch Up Pigment 37 Leather Refinishing 37 Base Coat 37 NOTE: COLORS ARE APPROXIMATE REPRESENTATIONS OF ACTUAL COLORS USING MODERN PROCESS TECHNIQUES.