Castanea Mollissima Blume): the Roots of Nut Tree Domestication
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

CHESTNUT (CASTANEA Spp.) CULTIVAR EVALUATION for COMMERCIAL CHESTNUT PRODUCTION
CHESTNUT (CASTANEA spp.) CULTIVAR EVALUATION FOR COMMERCIAL CHESTNUT PRODUCTION IN HAMILTON COUNTY, TENNESSEE By Ana Maria Metaxas Approved: James Hill Craddock Jennifer Boyd Professor of Biological Sciences Assistant Professor of Biological and Environmental Sciences (Director of Thesis) (Committee Member) Gregory Reighard Jeffery Elwell Professor of Horticulture Dean, College of Arts and Sciences (Committee Member) A. Jerald Ainsworth Dean of the Graduate School CHESTNUT (CASTANEA spp.) CULTIVAR EVALUATION FOR COMMERCIAL CHESTNUT PRODUCTION IN HAMILTON COUNTY, TENNESSEE by Ana Maria Metaxas A Thesis Submitted to the Faculty of the University of Tennessee at Chattanooga in Partial Fulfillment of the Requirements for the Degree of Master of Science in Environmental Science May 2013 ii ABSTRACT Chestnut cultivars were evaluated for their commercial applicability under the environmental conditions in Hamilton County, TN at 35°13ꞌ 45ꞌꞌ N 85° 00ꞌ 03.97ꞌꞌ W elevation 230 meters. In 2003 and 2004, 534 trees were planted, representing 64 different cultivars, varieties, and species. Twenty trees from each of 20 different cultivars were planted as five-tree plots in a randomized complete block design in four blocks of 100 trees each, amounting to 400 trees. The remaining 44 chestnut cultivars, varieties, and species served as a germplasm collection. These were planted in guard rows surrounding the four blocks in completely randomized, single-tree plots. In the analysis, we investigated our collection predominantly with the aim to: 1) discover the degree of acclimation of grower- recommended cultivars to southeastern Tennessee climatic conditions and 2) ascertain the cultivars’ ability to survive in the area with Cryphonectria parasitica and other chestnut diseases and pests present. -

Frank Meyer, Isabel Shipley Cunningham Agricultural Explorer
Frank Meyer, Isabel Shipley Cunningham Agricultural Explorer For 60 years the work of Frank N. Meyer has 2,500 pages of his letters tell of his journeys remained a neglected segment of America’s and the plants he collected, and the USDA heritage. Now, as people are becoming con- Inventory of Seeds and Plants Imported con- cerned about feeding the world’s growing tains descriptions of his introductions. population and about the loss of genetic di- Until recently little was known about the versity of crops, Meyer’s accomplishments first 25 years of Meyer’s life, when he lived have a special relevance. Entering China in in Amsterdam and was called Frans Meijer. 1905, near the dawn of the single era when Dutch sources reveal that he was bom into a explorers could travel freely there, he be- loving family in 1875. Frans was a quiet boy, came the first plant hunter to represent a who enjoyed taking long walks, reading government and to search primarily for eco- about distant lands, and working in his fami- nomically useful plants rather than orna- ly’s small garden. By the time he had mentals. No one before him had spent 10 fimshed elementary school, he knew that ’ years crossing the mountains, deserts, farms, wanted to be a world traveler who studied he ’,if and forests of Asia in search of fruits, nuts, plants; however, his parents could not afford vegetables, grains, and fodder crops; no one to give him further education. When he was has done so since. 14 years old, he found work as a gardener’s During four plant-hunting expeditions to helper at the Amsterdam Botanical Garden. -
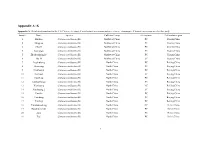
Appendix A~K
Appendix A~K Appendix A. Detailed information for the 146 Chinese chestnut (C.mollissima) accessions and nine chinese chinquapin (C.henryi) accessions used in this study Sample Name Species Cultivars Group Germplasm Cultivation region 1 Huijian Castanea mollissima Bl. Northwest China PC Shanxi,China 2 Mingjian Castanea mollissima Bl. Northwest China PC Shanxi,China 3 Chunli Castanea mollissima Bl. Northwest China PC Shanxi,China 4 Zuohongli Castanea mollissima Bl. Northwest China PC Shanxi,China 5 Zhenbashuangjie Castanea mollissima Bl. Northwest China PC Shanxi,China 6 zha 18 Castanea mollissima Bl. Northwest China LC Shanxi,China 7 Jingshuhong Castanea mollissima Bl. North China PC Beijing,China 8 Huaixiang Castanea mollissima Bl. North China PC Beijing,China 9 Huaihuang Castanea mollissima Bl. North China PC Beijing,China 10 Huzhaoli Castanea mollissima Bl. North China LC Beijing,China 11 Huaifeng Castanea mollissima Bl. North China PC Beijing,China 12 Yanshanhongli Castanea mollissima Bl. North China PC Beijing,China 13 Xiazhuang Castanea mollissima Bl. North China LC Beijing,China 14 Xinzhuang 2 Castanea mollissima Bl. North China LC Beijing,China 15 Huaijiu Castanea mollissima Bl. North China PC Beijing,China 16 Yanchang Castanea mollissima Bl. North China PC Beijing,China 17 Yanfeng Castanea mollissima Bl. North China PC Beijing,China 18 Yanshanzaofeng Castanea mollissima Bl. North China NC Hebei,China 19 Donglimingzhu Castanea mollissima Bl. North China PC Hebei,China 20 Zipo Castanea mollissima Bl. North China PC Hebei,China 21 Tasi Castanea mollissima Bl. North China PC Hebei,China 1 22 Zundali Castanea mollissima Bl. North China PC Hebei,China 23 Duanzhiyabian Castanea mollissima Bl. -

Castanea Mollissima
Castanea mollissima As the American chestnut struggles with disease, the blight- resistant Chinese chestnut is quickly gaining popularity. The sweet-tasting nuts are often roasted for holiday eating and have been made famous in turkey stuffing recipes across the country. But this is more than a nut tree. The shade of its spreading canopy is dense, providing relief in the hot, dry climates the Chinese chestnut does well in. Hardiness Zones: The chinese chestnut can be expected to grow in Hardiness Zones 4-8. Tree Type: This is a nut-producing tree, yielding nuts for human and wildlife consumption. Mature Size: The Chinese chestnut grows to a height of 40-60' and a spread of 40-60' at maturity. Growth Rate: This tree grows at a slow to medium rate, with height increases of anywhere from less than 12" to 24" per year. Sun Preference: Full sun is the ideal condition for this tree, meaning it should get at least six hours of direct, unfiltered sunlight each day. Soil Preference: The Chinese chestnut grows in acidic, loamy, moist, sandy, well-drained, and clay soils. It is drought-tolerant. Attributes This tree: Should be planted in pairs or groups to ensure pollination. 1 Yields a ripened nut crop mid/late September through October. A prickly 2-3 /2" seed husk en- closes 1-4 nuts. The nuts are large, meaty, crisp, and sweet, although less sweet than American chestnuts. Begins to bear nuts in 4-5 years if grown from seed. Provides dense shade with a handsome, spreading canopy. Has wood that is very durable and resistant to rot. -

Chestnut Growers Urged to Implement Quarantine for Chestnut Gall Wasp
Vol. 18 No. 4 Published by Chestnut Growers of America, Inc. Fall 2017 Chestnut Growers Urged to Implement Quarantine for Chestnut Gall Wasp By Michelle Warmund, Ph.D., University of Missouri Center for Agroforestry; Tom Green, Ph.D., Professor Emeritus, Western Illinois University; Tom Wahl, Red Fern Farms; Kathy Dice, Red Fern Farms; and Jim Dallmeyer, Thistle Creek Orchard he chestnut gall wasp, Dryocosumus 40 days and the larvae remain dormant Indeed, this pest was first introduced to Tkuriphilus Yasumatsu, is a tiny, gnat- until the following spring, when galls are the US on scion wood. Dispersal by flight sized, non-stinging insect that causes formed. With bud break, larvae induce is eclipsed by human transport. A serious galls in chestnut trees. These galls retard gall formation on developing plant tissues. source of propagation comes from home plant growth and flowering and can kill Larvae feed on the inner gall tissue for 20 owners who plant chestnuts in their yards branches. Severe infestations can kill trees. to 30 days before pupating. Adult wasps and hunters who plant them in woods to After the adult insects emerge, the dried, emerge from the galls in late May and attract deer. While commercial orchards blackened galls become woody and can early June. Beyond the gall clusters of dead may be fairly far apart, these alternate persist on older limbs for several years. leaves form. Called flags, these are easily growers provide additional “stepping Older, slower growing trees are more visible, making location of galls quickly stones” for the spread of the CGW. vulnerable. -

CHESTNUT CULTURE in CALIFORNIA UC Dept
CHESTNUT CULTURE in CALIFORNIA UC Dept. of Ag & Natl. Resources Publication # 8010 - By Paul Vossen The chestnut is a delicious nut produced on large magnificent trees on millions of acres of native habitat in the Northern Hemisphere, particularly China, Korea, Japan, and Southern Europe. The entire Eastern half of the USA was once covered with native chestnut trees until a blight fungus introduced from Asia destroyed them in the early 1900’s. The fleshy nut is sweet with a starchy texture and a low fat content resembling a cereal grain. The nuts are eaten as traditional foods in much of Asia and Europe where they are consumed fresh, cooked, candied, and as a source of flour for pastries. The chestnut tree (Castanea sp.) is in the same family as the beeches and oaks (Fagaceae). The formidable, spiny chestnut burr is the equivalent to the cap on an acorn. Chestnuts belong to the Genus: Castanea, with four main economic species: C. dentata (North American), C. mollissima (Chinese), C. sativa (European), and C. crenata (Japanese). It is not related to the horse chestnut (Aesculus sp.). The tree has a gray bark and is deciduous with leaves 5-7 inches long, sharply serrated, oblong-lanceolate, and pinnately veined. Domestication of the chestnut is still progressing with much of the world’s production collected from natural stands. SPECIES Four species of chestnut are grown in North America. They exist as pure species or, more commonly, as hybrids of the various species because they readily cross with one another. In many cases, it is difficult to distinguish species and almost impossible to determine the parentage of the hybrids visually. -

Pollen Morphology of Chestnut Cultivars in Southern China by Using Scanning Electron Microscope
Bangladesh J. Bot. 49(1): 39-46, 2020 (March) POLLEN MORPHOLOGY OF CHESTNUT CULTIVARS IN SOUTHERN CHINA BY USING SCANNING ELECTRON MICROSCOPE HUAN XIONG, DE-YI YUAN, YA CHEN, GENHUA NIU AND FENG ZOU* The Key Lab of Non-wood Forest Products of Forestry Ministry , Central South University of Forestry and Technology, Changsha 410004, Hunan, P. R. China Keywords: Pollen morphology, Chestnut, Pollen axis length, Equatorial width, Colpus length Abstract The morphological characteristics of pollen grains of 16 cultivars belonging to two species of the genus Castanea (Castanea mollissima and C. henryi) collected from southern China were observed by using scanning electron microscope (SEM). The length of polar axis (PL), the equatorial diameter (ED), the length of colpus (CL) of grain, P/E ratio (polar axis to equatorial diameter), P/C ratio (polar axis to colpus length), and C/E ratio (colpus length to equatorial diameter) were measured and their variations were compared among studied taxa. The result of this study indicated that chestnut pollen grain was a tricolporate in aperture type, and aperprolate type in equatorial view. The morphological characteristics were mainly represented by PL, ED and CL as revealed by principal components analysis, which accounted for 99.56% of the total variations. The cluster analysis by the UPGMA tree divided the taxa into three groups and showed a great morphological affinity among the 16 cultivars. Introduction Chestnut has been placed in the family of Fagaceae which is monoecious, wind-pollinated and self-incompatible (Mckay 1942, Bounous and Marinoni 2005). In China, this species is economically important for timber and nut production. -

Restoration of the American Chestnut in New Jersey
U.S. Fish & Wildlife Service Restoration of the American Chestnut in New Jersey The American chestnut (Castanea dentata) is a tree native to New Jersey that once grew from Maine to Mississippi and as far west as Indiana and Tennessee. This tree with wide-spreading branches and a deep broad-rounded crown can live 500-800 years and reach a height of 100 feet and a diameter of more than 10 feet. Once estimated at 4 billion trees, the American chestnut Harvested chestnuts, early 1900's. has almost been extirpated in the last 100 years. The U.S. Fish and Wildlife Service, New Jersey Field Value Office (Service) and its partners, including American Chestnut The American chestnut is valued Cooperators’ Foundation, American for its fruit and lumber. Chestnuts Chestnut Foundation, Monmouth are referred to as the “bread County Parks, Bayside State tree” because their nuts are Prison, Natural Lands Trust, and so high in starch that they can several volunteers, are working to American chestnut leaf (4"-8"). be milled into flour. Chestnuts recover the American chestnut in can be roasted, boiled, dried, or New Jersey. History candied. The nuts that fell to the ground were an important cash Chestnuts have a long history of crop for families in the northeast cultivation and use. The European U.S. and southern Appalachians chestnut (Castanea sativa) formed up until the twentieth century. the basis of a vital economy in Chestnuts were taken into towns the Mediterranean Basin during by wagonload and then shipped Roman times. More recently, by train to major markets in New areas in Southern Europe (such as York, Boston, and Philadelphia. -

How a Flower Becomes a Chestnut: Morphological Development of Chinese Chestnuts (Castanea Mollisima)
How a Flower Becomes a Chestnut: Morphological Development of Chinese Chestnuts (Castanea mollisima) Amy Miller1, Diane D, Miller2, and Paula M. Pijut3 1Department of Forestry and Natural Resources Hardwood Tree Inprovement and Regeneration Center Purdue University 715 W. State St. West Lafayette, IN 47907 2Ohio Agricultural Research and Development Center The Ohio State University Wooster, OH 44691 3Northern Research Station Hardwood Tree Inprovement and Regeneration Center USDA Forest Service West Lafayette, IN 47907 [email protected] Ph: (330) 413-9384, Fax: (765) 494-9461 hestnuts, members of the genus Castanea, family (Anagnostakis 1987). Efforts are ongoing to produce and CFagaceae, are popular worldwide and consist of three introduce blight-resistant, well-adapted chestnut back to the sections with at least seven distinct species, but may include North American forest to regain its ecological and economic up to 12 species according to their classification (Bounous benefits (Thompson 2012). and Marinoni 2005). All species have noteworthy ecologi- Of the seven distinct species, three chestnut species, cal, economic, and cultural importance in southern Europe, Chinese chestnut (C. mollissima Blume), Chinese chinqua- Anatolia, the Caucasus Mountains, temperate eastern Asia, pin (C. henryi (Skan.) Rehder. and E.H. Wilson), and Seguin and eastern North America (Conedera et al. 2004; Davis chestnut (C. seguinii Dode.) are native to China, Japanese 2006). Chestnut species regularly bear sweet, nutritious chestnut (C. crenata Siebold and Zucc.) is native to Japan nuts that are high in carbohydrate, but low in fat (Bounous and Korea, European or Sweet chestnut (C. sativa Mill.) is and Marinoni 2005; McCarthy and Meredith 1988; Senter found in Europe, Anatolia, and the Caucasus, and American et al. -

Asia 24 Supplementary Material
Asia 24 Supplementary Material Coordinating Lead Authors: Yasuaki Hijioka (Japan), Erda Lin (China), Joy Jacqueline Pereira (Malaysia) Lead Authors: Richard T. Corlett (China), Xuefeng Cui (China), Gregory Insarov (Russian Federation), Rodel Lasco (Philippines), Elisabet Lindgren (Sweden), Akhilesh Surjan (India) Contributing Authors: Elena M. Aizen (USA), Vladimir B. Aizen (USA), Rawshan Ara Begum (Bangladesh), Kenshi Baba (Japan), Monalisa Chatterjee (USA/India), J. Graham Cogley (Canada), Noah Diffenbaugh (USA), Li Ding (Singapore), Qingxian Gao (China), Matthias Garschagen (Germany), Masahiro Hashizume (Japan), Manmohan Kapshe (India), Andrey G. Kostianoy (Russia), Kathleen McInnes (Australia), Sreeja Nair (India), S.V.R.K. Prabhakar (India), Yoshiki Saito (Japan), Andreas Schaffer (Singapore), Rajib Shaw (Japan), Dáithí Stone (Canada/South Africa /USA), Reiner Wassman (Philippines), Thomas J. Wilbanks (USA), Shaohong Wu (China) Review Editors: Rosa Perez (Philippines), Kazuhiko Takeuchi (Japan) Volunteer Chapter Scientists: Yuko Onishi (Japan), Wen Wang (China) This chapter on-line supplementary material should be cited as: Hijioka , Y., E. Lin, J.J. Pereira, R.T. Corlett, X. Cui, G.E. Insarov, R.D. Lasco, E. Lindgren, and A. Surjan, 2014: Asia – supplementary material. In: Climate Change 2014: Impacts, Adaptation, and Vulnerability. Part B: Regional Aspects. Contribution of Working Group II to the Fifth Assessment Report of the Intergovernmen tal Panel on Climate Change [Barros, V.R., C.B. Field, D.J. Dokken, M.D. Mastrandrea, K.J. Mach, T.E. Bilir, M. Chatterjee, K.L. Ebi, Y.O. Estrada, R.C. Genova, B. Girma, E.S. Kissel, A.N. Levy, S. MacCracken, P.R. Mastrandrea, and L.L. White (eds.)]. Available from www.ipcc-wg2.gov/AR5 and www.ipcc.ch. -

Comparative Study on Phytochemical Profiles and Antioxidant Capacities
antioxidants Article Comparative Study on Phytochemical Profiles and Antioxidant Capacities of Chestnuts Produced in Different Geographic Area in China 1,2, 1, 1, 1, Ziyun Xu y, Maninder Meenu y, Pengyu Chen y and Baojun Xu * 1 Food Science and Technology Programme, Beijing Normal University-Hong Kong Baptist University United International College, Zhuhai 519087, China; [email protected] (Z.X.); [email protected] (M.M.); [email protected] (P.C.) 2 Department of Food Science and Agricultural Chemistry, McGill University, Quebec, QC H9X 3V9, Canada * Correspondence: [email protected]; Tel.: +86-7563620636; Fax: +86-7563620882 These authors contributed equally to the article as the first authors. y Received: 27 January 2020; Accepted: 22 February 2020; Published: 25 February 2020 Abstract: This study aimed to systematically assess the phenolic profiles and antioxidant capacities of 21 chestnut samples collected from six geographical areas of China. All these samples exhibit significant differences (p < 0.05) in total phenolic contents (TPC), total flavonoids content (TFC), condensed tannin content (CTC) and antioxidant capacities assessed by DPPH free radical scavenging capacity (DPPH), ABTS free radical scavenging capacities (ABTS), ferric reducing antioxidant power (FRAP), and 14 free phenolic acids. Chestnuts collected from Fuzhou, Jiangxi (East China) exhibited the maximum values for TPC (2.35 mg GAE/g), CTC (13.52 mg CAE/g), DPPH (16.74 µmol TE/g), ABTS (24.83 µmol TE/g), FRAP assays (3.20 mmol FE/100 g), and total free phenolic acids (314.87 µg/g). Vanillin and gallic acids were found to be the most abundant free phenolic compounds among other 14 phenolic compounds detected by HPLC. -

Chestnuts Bred for Blight Resistance Depart Nursery with Distinct Fungal Rhizobiomes
Mycorrhiza (2019) 29:313–324 https://doi.org/10.1007/s00572-019-00897-z ORIGINAL ARTICLE Chestnuts bred for blight resistance depart nursery with distinct fungal rhizobiomes Christopher Reazin1 & Richard Baird2 & Stacy Clark3 & Ari Jumpponen1 Received: 15 January 2019 /Accepted: 9 May 2019 /Published online: 25 May 2019 # Springer-Verlag GmbH Germany, part of Springer Nature 2019 Abstract Restoration of the American chestnut (Castanea dentata) is underway using backcross breeding that confers chestnut blight disease resistance from Asian chestnuts (most often Castanea mollissima) to the susceptible host. Successful restoration will depend on blight resistance and performance of hybrid seedlings, which can be impacted by below-ground fungal communities. We compared fungal communities in roots and rhizospheres (rhizobiomes) of nursery-grown, 1-year-old chestnut seedlings from different genetic families of American chestnut, Chinese chestnut, and hybrids from backcross breeding generations as well as those present in the nursery soil. We specifically focused on the ectomycorrhizal (EcM) fungi that may facilitate host performance in the nursery and aid in seedling establishment after outplanting. Seedling rhizobiomes and nursery soil communities were distinct and seedlings recruited heterogeneous communities from shared nursery soil. The rhizobiomes included EcM fungi as well as endophytes, putative pathogens, and likely saprobes, but their relative proportions varied widely within and among the chestnut families. Notably, hybrid seedlings that hosted few EcM fungi hosted a large proportion of potential pathogens and endophytes, with possible consequences in outplanting success. Our data show that chestnut seedlings recruit divergent rhizobiomes and depart nurseries with communities that may facilitate or compromise the seedling performance in the field.