Print This Article
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Download PDF (English)
Revista Brasileira de Farmacognosia 26 (2016) 619–626 ww w.elsevier.com/locate/bjp Original Article Evaluation of mechanism for antihypertensive and vasorelaxant effects of hexanic and hydroalcoholic extracts of celery seed in normotensive and hypertensive rats a b c Faezeh Tashakori-Sabzevar , Bibi Marjan Razavi , Mohsen Imenshahidi , a a a Mohammadreza Daneshmandi , Hamidreza Fatehi , Yaser Entezari Sarkarizi , c,∗ Seyed Ahmad Mohajeri a Student Research Committee, Mashhad University of Medical Sciences, Mashhad, Iran b Targeted Drug Delivery Research Center, School of Pharmacy, Mashhad University of Medical Sciences, Mashhad, Iran c Pharmaceutical Research Center, School of Pharmacy, Mashhad University of Medical Sciences, Mashhad, Iran a b s t r a c t a r t i c l e i n f o Article history: Celery (Apium graveolens L., Apiaceae) is one of the popular aromatic vegetables and part of the daily diet Received 26 December 2015 around the world. In this study, aqueous-ethanolic and hexane extracts of celery seed were prepared and Accepted 23 May 2016 the amount of n-butylphthalide, as an active component, was determined in each extract. Then the effects Available online 26 June 2016 of hexanic extract on systolic, diastolic, mean arterial blood pressure and heart rate were evaluated in an invasive rat model. The vasodilatory effect and possible mechanisms of above mentioned extracts on aorta Keywords: ring were also measured. High performance liquid chromatography analysis revealed that hexanic extract Celery seed contains significantly higher amounts of n-butylphthalide, compared to aqueous-ethanolic extract. The Heart rate results indicated that hexanic extract significantly decreased the systolic, diastolic, mean arterial blood Hypotensive effects pressure and heart rate in normotensive and hypertensive rats. -

Effect of Angelica Archangelica L. Extract on Growth Performance
232 Bulgarian Journal of Agricultural Science, 26 (No 1) 2020, 232–237 Effect ofAngelica archangelica L. extract on growth performance, meat quality and biochemical blood parameters of rainbow trout (Oncorhynchus mykiss W.), cultivated in a recirculating system Radoslav Koshinski, Katya Velichkova*, Ivaylo Sirakov and Stefka Stoyanova Trakia University, Department of Biology and Aquaculture, Faculty of Agriculture, 6014 Stara Zagora, Bulgaria *Corresponding author: [email protected] Abstract Koshinski, R., Velichkova, K., Sirakov, I. & Stoyanova, St. (2020). Effect of Angelica archangelica L. extract on growth performance, meat quality and biochemical blood parameters of rainbow trout (Oncorhynchus mykiss W.), cultivated in a recirculating system. Bulg. J. Agric. Sci., 26 (1), 232–237 The medicinal herbs as natural products can be use like not expensive additives in artificial diets for aquatic animals which are safe for fish and the environment. The purpose of this study is to determine the effect of the Angelica archangelica L. extract on the growth performance, meat quality and biochemical blood parameters (glucose, urea, creatinine, total protein, albumin, ASAT, ALAT, ALP, Ca, P, Mg, triglycerides, cholesterol) of rainbow trout (Oncorhynchus mykiss). Thirty specimens from the rainbow trout with anaverage weight of 42.55±7.48 g (control, C) and 42.51±6.02 g (experimental, Ang.a.) in good health condition were placed in each tank and cultivated for 60 days. A control group (no added) and an experimental (with added 433 mg.kg-1 of angelica extract) option, each with a two repetition, were set in a recirculating system in the Aquaculture Base of the Faculty of Agriculture at the Trakia University. -

Research Article Nutrition in Herbal Plants Used in Saudi Arabia
Hindawi Scientifica Volume 2020, Article ID 6825074, 9 pages https://doi.org/10.1155/2020/6825074 Research Article Nutrition in Herbal Plants Used in Saudi Arabia Hanan Almahasheer Department of Biology, College of Science, Imam Abdulrahman Bin Faisal University (IAU), Dammam 31441-1982, Saudi Arabia Correspondence should be addressed to Hanan Almahasheer; [email protected] Received 4 October 2019; Accepted 30 March 2020; Published 28 April 2020 Academic Editor: Roland Bitsch Copyright © 2020 Hanan Almahasheer. 'is is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Herbs are used for centuries by many people worldwide. 'is study derives insights into the use and content of herbs that are consumed among Saudi citizens. An online questionnaire was distributed to understand the basic information about Saudi citizens’ preference and daily patterns of herbal plants that are usually used as drinks. Moreover, concentrations of fourteen elements in twenty-one herbal plants that were indicated in the previous questionnaire were collected from the local market and then analyzed using an Inductively Coupled Plasma Emission Spectrometry (ICP). Mint leaves were significantly higher in most of the nutrients analyzed, and mint was the most popular drink among participants, followed by green tea and anise. Most of the citizens preferred to drink one cup only at home and believed that herbs are good for their health and potentially could help them to sleep better. 'e outcomes derived from this research could help future assessments of diet patterns among Saudi citizens. -
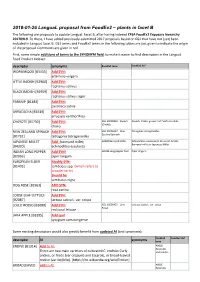
2018-01-26 Langual Proposal from Foodex2 – Plants in Facet B
2018-01-26 LanguaL proposal from FoodEx2 – plants in facet B The following are proposals to update LanguaL Facet B, after having indexed EFSA FoodEx2 Exposure hierarchy 20170919. To these, I have added previously-submitted 2017 proposals based on GS1 that have not (yet) been included in LanguaL facet B. GS1 terms and FoodEx2 terms in the following tables are just given to indicate the origin of the proposal. Comments are given in red. First, some simple additions of terms to the SYNONYM field, to make it easier to find descriptors in the LanguaL Food Product Indexer: descriptor synonyms FoodEx2 term FoodEx2 def WORMWOOD [B3433] Add SYN: artemisia vulgaris LITTLE RADISH [B2960] Add SYN: raphanus sativus BLACK RADISH [B2959] Add SYN: raphanus sativus niger PARSNIP [B1483] Add SYN: pastinaca sativa ARRACACHA [B3439] Add SYN: arracacia xanthorrhiza CHAYOTE [B1730] Add SYN: GS1 10006356 - Squash Squash, Choko, grown from Sechium edule (Choko) choko NEW ZEALAND SPINACH Add SYN: GS1 10006427 - New- Tetragonia tetragonoides Zealand Spinach [B1732] tetragonia tetragonoides JAPANESE MILLET Add : barnyard millet; A000Z Barnyard millet Echinochloa esculenta (A. Braun) H. Scholz, Barnyard millet or Japanese Millet. [B4320] echinochloa esculenta INDIAN LONG PEPPER Add SYN! A019B Long pepper fruit Piper longum [B2956] piper longum EUROPEAN ELDER Modify SYN: [B1403] sambucus spp. (which refers to broader term) Should be sambucus nigra DOG ROSE [B2961] ADD SYN: rosa canina LOOSE LEAF LETTUCE Add SYN: [B2087] lactusa sativa L. var. crispa LOLLO ROSSO [B2088] Add SYN: GS1 10006425 - Lollo Lactuca sativa L. var. crispa Rosso red coral lettuce JAVA APPLE [B3395] Add syn! syzygium samarangense Some existing descriptors would also greatly benefit from updated AI (and synonyms): FoodEx2 FoodEx2 def descriptor AI synonyms term ENDIVE [B1314] Add to AI: A00LD Escaroles There are two main varieties of cultivated C. -

Periodic Table of Herbs 'N Spices
Periodic Table of Herbs 'N Spices 11HH 1 H 2 HeHe Element Proton Element Symbol Number Chaste Tree Chile (Vitex agnus-castus) (Capsicum frutescens et al.) Hemptree, Agnus Cayenne pepper, Chili castus, Abraham's balm 118Uuo Red pepper 33LiLi 44 Be 5 B B 66 C 7 N 7N 88O O 99 F 1010 Ne Ne Picture Bear’s Garlic Boldo leaves Ceylon Cinnamon Oregano Lime (Allium ursinum) (Peumus boldus) (Cinnamomum zeylanicum) Nutmeg Origanum vulgare Fenugreek Lemon (Citrus aurantifolia) Ramson, Wild garlic Boldina, Baldina Sri Lanka cinnamon (Myristica fragrans) Oregan, Wild marjoram (Trigonella foenum-graecum) (Citrus limon) 11 Na Na 1212 Mg Mg 1313 Al Al 1414 Si Si 1515 P P 16 S S 1717 Cl Cl 1818 Ar Ar Common Name Scientific Name Nasturtium Alternate name(s) Allspice Sichuan Pepper et al. Grains of Paradise (Tropaeolum majus) (Pimenta dioica) (Zanthoxylum spp.) Perilla (Aframomum melegueta) Common nasturtium, Jamaica pepper, Myrtle Anise pepper, Chinese (Perilla frutescens) Guinea grains, Garden nasturtium, Mugwort pepper, Pimento, pepper, Japanese Beefsteak plant, Chinese Savory Cloves Melegueta pepper, Indian cress, Nasturtium (Artemisia vulgaris) Newspice pepper, et al. Basil, Wild sesame (Satureja hortensis) (Syzygium aromaticum) Alligator pepper 1919 K K 20 Ca Ca 2121 Sc Sc 2222 Ti Ti 23 V V 24 Cr Cr 2525 Mn Mn 2626 Fe Fe 2727 Co Co 2828 Ni Ni 29 Cu Cu 3030 Zn Zn 31 Ga Ga 3232 Ge Ge 3333As As 34 Se Se 3535 Br Br 36 Kr Kr Cassia Paprika Caraway (Cinnamomum cassia) Asafetida Coriander Nigella Cumin Gale Borage Kaffir Lime (Capsicum annuum) (Carum carvi) -

Petiole Usually Stipules at the Base
Umbelliferae P. Buwalda Groningen) Annual or perennial herbs, never woody shrubs (in Malaysia). Stems often fur- rowed and with soft pith. Leaves alternate along the stems, often also in rosettes; petiole usually with a sheath, sometimes with stipules at the base; lamina usually much divided, sometimes entire. Flowers polygamous, in simple or compound um- bels, sometimes in heads, terminal or leaf-opposed, beneath with or without in- volucres and involucels. Calyx teeth 5, often obsolete. Petals 5, alternate with the calyx teeth, equal or outer ones of the inflorescence enlarged, entire or more or less divided, often with inflexed tips, inserted below the epigynous disk. Stamens alter- nate with the petals, similarly inserted. Disk 2-lobed, free from the styles or con- fluent with their thickened base, forming a stylopodium. Ovary inferior; styles 2. Fruits with 2 one-seeded mericarps, connected by a narrow or broad junction (commissure) in fruit separating, leaving sometimes a persistent axis (carpophore) either entire or splitting into 2 halves; mericarps with 5 longitudinal ribs, 1 dorsal rib at the back of the mericarp, 2 lateral ribs at the commissure; 2 intermediate ribs between the dorsal and the lateral ones; sometimes with secondary ribs be- without fascicular often vittae in tween the primary ones, these bundles; the ridges between the ribs or under the secondary ribs, and in the commissure, seldom under the primary ribs. Numerous Distr. genera and species, all over the world. The representatives native in Malaysia belong geographically to five types. (1) Ubiquitous genera (Hydrocotyle, Centella, Oenanthe); one species, & Hydrocotyle vulgaris, shows a remarkable disjunction, occurring in Europe N. -

INDEX for 2011 HERBALPEDIA Abelmoschus Moschatus—Ambrette Seed Abies Alba—Fir, Silver Abies Balsamea—Fir, Balsam Abies
INDEX FOR 2011 HERBALPEDIA Acer palmatum—Maple, Japanese Acer pensylvanicum- Moosewood Acer rubrum—Maple, Red Abelmoschus moschatus—Ambrette seed Acer saccharinum—Maple, Silver Abies alba—Fir, Silver Acer spicatum—Maple, Mountain Abies balsamea—Fir, Balsam Acer tataricum—Maple, Tatarian Abies cephalonica—Fir, Greek Achillea ageratum—Yarrow, Sweet Abies fraseri—Fir, Fraser Achillea coarctata—Yarrow, Yellow Abies magnifica—Fir, California Red Achillea millefolium--Yarrow Abies mariana – Spruce, Black Achillea erba-rotta moschata—Yarrow, Musk Abies religiosa—Fir, Sacred Achillea moschata—Yarrow, Musk Abies sachalinensis—Fir, Japanese Achillea ptarmica - Sneezewort Abies spectabilis—Fir, Himalayan Achyranthes aspera—Devil’s Horsewhip Abronia fragrans – Sand Verbena Achyranthes bidentata-- Huai Niu Xi Abronia latifolia –Sand Verbena, Yellow Achyrocline satureoides--Macela Abrus precatorius--Jequirity Acinos alpinus – Calamint, Mountain Abutilon indicum----Mallow, Indian Acinos arvensis – Basil Thyme Abutilon trisulcatum- Mallow, Anglestem Aconitum carmichaeli—Monkshood, Azure Indian Aconitum delphinifolium—Monkshood, Acacia aneura--Mulga Larkspur Leaf Acacia arabica—Acacia Bark Aconitum falconeri—Aconite, Indian Acacia armata –Kangaroo Thorn Aconitum heterophyllum—Indian Atees Acacia catechu—Black Catechu Aconitum napellus—Aconite Acacia caven –Roman Cassie Aconitum uncinatum - Monkshood Acacia cornigera--Cockspur Aconitum vulparia - Wolfsbane Acacia dealbata--Mimosa Acorus americanus--Calamus Acacia decurrens—Acacia Bark Acorus calamus--Calamus -

Caraway As Important Medicinal Plants in Management of Diseases
Natural Products and Bioprospecting https://doi.org/10.1007/s13659-018-0190-x (012 3456789().,- volV)(0123456789().,-volV) REVIEW Caraway as Important Medicinal Plants in Management of Diseases Mohaddese Mahboubi1 Received: 2 August 2018 / Accepted: 19 October 2018 Ó The Author(s) 2018 Abstract Carum carvi or caraway is traditionally used for treatment of indigestion, pneumonia, and as appetizer, galactagogue, and carminative. Essential oil, fixed oil and many other valuable extractive compounds with industrial applications are prepared from caraway. This review article has new deep research on caraway as medicinal plant. For preparing the manuscript, the information was extracted from accessible international databases (Google scholar, PubMed, Science direct, Springer, and Wiley), electronic resources and traditional books by key word of caraway or Carum carvi. The results of traditional studies exhibited that the galactagogue and carminative effects of caraway fruits are superior to other effects. Although, the traditional scholars used it as appetizer, while caraway was the main ingredient of anti-obesity drugs in traditional medicine, which has been confirmed in two modern clinical trials of human studies. Caraway oil in combination with peppermint oil or menthol is used for treatment of functional dyspepsia in clinical studies. Caraway oil topically on abdomen relieves the IBS symptoms in patient. Although, the use of caraway oil is not recommended in adults under 18 years due to insufficient data, but it can topically use as anti-colic and carminative agent in children or infants. The anti- aflatoxigenic, antioxidant and antimicrobial effects of caraway oil along with its reputation as spice help the industries to use it as natural preservatives and antioxidant agents. -

Antioxidant and Antibacterial Investigations on Essential Oils And
Natural Product Radiance, Vol. 6(2), 2007, pp.114-121 Research Article Antioxidant and antibacterial investigations on essential oils and acetone extracts of some spicesÜ Gurdip Singh1*, Sumitra Maurya3, Palanisamy Marimuthu4, H S Murali2 and A S Bawa2 1Chemistry Department, DDU Gorakhpur University, Gorakhpur- 273 009, Uttar Pradesh, India 2Defence Food Research Laboratory, Mysore- 570 011, Karnataka, India 3 Agharkar Research Institute, Pune-411 004, Maharashtra, India 4 Indian Agricultural Research Institute, New Delhi- 110 012, India †Part 49 *Correspondent author, E-mail: [email protected] Phone: 91-551-2200745 (R) 2202856 (O); Fax: 91-551-2340459; Received 7 April 2006; Accepted 11 September 2006 Abstract The studies on antioxidant and antibacterial potential of essential/volatile oils and acetone extracts of various spices are presented in this paper. The antibacterial activity of the volatile oils and acetone extracts of anise, ajwain, tejpat, Chinese Cassia bark, fennel, coriander, dill, turmeric and star anise have been studied against Escherichia coli, Salmonella typhi, Pseudomonas aeruginosa, Bacillus cereus, B. subtilis and Staphylococcus aureus by disc diffusion and plate count methods. The results showed that volatile oils and extracts varied in their bioactivity. The volatile oils of ajwain, tejpat, Chinese Cassia bark and coriander were found to possess Ajwain excellent activity against all the Gram positive and Gram negative bacterial strains tested. These volatile oils and extracts are equally or more effective when compared with standard antibiotics diffusion and dilution of the test substance even at very low concentration. However, the acetone extract was found to be less effective as compared to volatile oils. Antioxidant activity of the oils and extracts were studied by DPPH, in a microbiological medium. -

Essential Oils and Uses of Eryngium Foetidum L
Essential oils and uses of Eryngium foetidum L. Marina Silalahi * Biology Education Department, FKIP, Universitas Kristen Indonesia, Jakarta. Jl. Mayjen Sutoyo No. 2 Cawang, Jakarta Timur. 1350. GSC Biological and Pharmaceutical Sciences, 2021, 15(03), 289–294 Publication history: Received on 17 May 2021; revised on 25 June 2021; accepted on 27 June 2021 Article DOI: https://doi.org/10.30574/gscbps.2021.15.3.0175 Abstract Eryngium foetidum is a species belonging Apiaceae which is used as medicine, vegetables, and spices. The plants used as medicine related to its secondary metabolites. The writing of this article is based on a literature review obtained online sources and offline used keywords Eryngium foetidum, secondary metabolites of E. foetidum, and uses of E. foetidum. In ethnobotany E. foetidum used to treat fever, hypertension, headache, abdominal pain, asthma, arthritis, diarrhea, and malaria. The essential oil of E. foetidum is dominated by (E) -2-dodecenal, dodecanoic acid, trans-2- dodecanoic acid, (E) -2-tridecenal, duraldehyde, and tetradecane. The bioactivity of E. foetidum has anti-microbial, antioxidant and anti-inflammatory. The ability of E. foetidum essential oils as an anti-microbial is very potential to be developed as a natural food preservative. Keywords: Eryngium foetidum; Anti-microbial; Essential oil; antioxidant 1. Introduction Plants as medicinal and food ingredients a very important in the development of human civilization. The plants are direct or indirect impact to human health, so it’s to be carried out as new alternative therapeutic agents. The plants more than 7,000 species used as traditional medicinal. The plants efficacy is related to their bioactive compounds such as phenols, flavonoids, and tannins [1]. -

Plants and Parts of Plants Used in Food Supplements: an Approach to Their
370 ANN IST SUPER SANITÀ 2010 | VOL. 46, NO. 4: 370-388 DOI: 10.4415/ANN_10_04_05 ES I Plants and parts of plants used OLOG D in food supplements: an approach ETHO to their safety assessment M (a) (b) (b) (b) D Brunella Carratù , Elena Federici , Francesca R. Gallo , Andrea Geraci , N (a) (b) (b) (a) A Marco Guidotti , Giuseppina Multari , Giovanna Palazzino and Elisabetta Sanzini (a)Dipartimento di Sanità Pubblica Veterinaria e Sicurezza Alimentare; RCH (b) A Dipartimento del Farmaco, Istituto Superiore di Sanità, Rome, Italy ESE R Summary. In Italy most herbal products are sold as food supplements and are subject only to food law. A list of about 1200 plants authorised for use in food supplements has been compiled by the Italian Ministry of Health. In order to review and possibly improve the Ministry’s list an ad hoc working group of Istituto Superiore di Sanità was requested to provide a technical and scientific opinion on plant safety. The listed plants were evaluated on the basis of their use in food, therapeu- tic activity, human toxicity and in no-alimentary fields. Toxicity was also assessed and plant limita- tions to use in food supplements were defined. Key words: food supplements, botanicals, herbal products, safety assessment. Riassunto (Piante o parti di piante usate negli integratori alimentari: un approccio per la valutazione della loro sicurezza d’uso). In Italia i prodotti a base di piante utilizzati a scopo salutistico sono in- tegratori alimentari e pertanto devono essere commercializzati secondo le normative degli alimenti. Le piante che possono essere impiegate sono raccolte in una “lista di piante ammesse” stabilita dal Ministero della Salute. -

Chemical Composition of Trachyspermum Ammi L. and Its
Journal of Pharmacognosy and Phytochemistry 2017; 6(3): 131-140 E-ISSN: 2278-4136 P-ISSN: 2349-8234 Chemical composition of Trachyspermum ammi L. and JPP 2017; 6(3): 131-140 Received: 04-03-2017 its biological properties: A review Accepted: 05-04-2017 KK Chahal KK Chahal, K Dhaiwal, A Kumar, D Kataria and N Singla Department of Chemistry, Punjab Agricultural University, Ludhiana, Punjab, India Abstract Trachyspermum ammi L. (Apiaceae) commonly known as ajwain is an important medicinal, aromatic K Dhaiwal and spice plant. It was originated in Egypt and widely distributed throughout the World. Ajwain seeds Department of Chemistry, yield 2-5% brownish essential oil, with thymol as the major constituent along with p-cymene, γ- Punjab Agricultural University, terpinene, α-pinene, β-pinene and α-terpinene. Due to presence of various chemical constituents in Ludhiana, Punjab, India ajwain, various biological and pharmacological properties have been reported. The present study is an effort to collect all the information regarding chemical composition and biological activities of ajwain. A Kumar Department of Chemistry, Punjab Agricultural University, Keywords: Trachyspermum ammi (L.), Essential oil, Chemical composition, Biological properties Ludhiana, Punjab, India 1. Introduction D Kataria Natural products, such as essential oils are produced by the secondary metabolism in plants. Department of Chemistry, Their constituents are used in human consumption as functional food, food additives, Punjab Agricultural University, Ludhiana, Punjab, India medicines, nutritional supplements and for the manufacture of cosmetics (Burt and Reinders [15] 2003) . The volatile components of essential oils mainly consist of monoterpenes, N Singla sesquiterpenes and their oxygenated derivatives such as alcohols, aldehyde, ketones, acids and Department of Chemistry, esters (Suntar et al.