Modelling the Solubility of the 20 Proteinogenic Amino Acids with Experimentally Derived Saturation Data
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Direct Sensing and Discrimination Among Ubiquitin and Ubiquitin Chains Using Solid-State Nanopores
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector 2340 Biophysical Journal Volume 108 May 2015 2340–2349 Article Direct Sensing and Discrimination among Ubiquitin and Ubiquitin Chains Using Solid-State Nanopores Iftach Nir,1 Diana Huttner,1 and Amit Meller1,* 1Department of Biomedical Engineering, The Technion—Israel Institute of Technology, Haifa, Israel ABSTRACT Nanopore sensing involves an electrophoretic transport of analytes through a nanoscale pore, permitting label- free sensing at the single-molecule level. However, to date, the detection of individual small proteins has been challenging, primarily due to the poor signal/noise ratio that these molecules produce during passage through the pore. Here, we show that fine adjustment of the buffer pH, close to the isoelectric point, can be used to slow down the translocation speed of the analytes, hence permitting sensing and characterization of small globular proteins. Ubiquitin (Ub) is a small protein of 8.5 kDa, which is well conserved in all eukaryotes. Ub conjugates to proteins as a posttranslational modification called ubiquiti- nation. The immense diversity of Ub substrates, as well as the complexity of Ub modification types and the numerous physio- logical consequences of these modifications, make Ub and Ub chains an interesting and challenging subject of study. The ability to detect Ub and to identify Ub linkage type at the single-molecule level may provide a novel tool for investigation in the Ub field. This is especially adequate because, for most ubiquitinated substrates, Ub modifies only a few molecules in the cell at a given time. -

Effect of Peptide Histidine Isoleucine on Water and Electrolyte Transport in the Human Jejunum
Gut: first published as 10.1136/gut.25.6.624 on 1 June 1984. Downloaded from Gut, 1984, 25, 624-628 Alimentary tract and pancreas Effect of peptide histidine isoleucine on water and electrolyte transport in the human jejunum K J MORIARTY, J E HEGARTY, K TATEMOTO, V MUTT, N D CHRISTOFIDES, S R BLOOM, AND J R WOOD From the Department of Gastroenterology, St Bartholomew's Hospital, London, The Liver Unit, King's College Hospital, London, Department ofMedicine, Hammersmith Hospital, London, and Department of Biochemistry, Karolinska Institute, Stockholm, Sweden SUMMARY Peptide histidine isoleucine, a 27 amino acid peptide with close amino acid sequence homology to vasoactive intestinal peptide and secretin, is distributed throughout the mammalian intestinal tract, where it has been localised to intramural neurones. An intestinal perfusion technique has been used to study the effect of intravenous peptide histidine isoleucine (44.5 pmol/kg/min) on water and electrolyte transport from a plasma like electrolyte solution in human jejunum in vivo. Peptide histidine isoleucine infusion produced peak plasma peptide histidine isoleucine concentrations in the range 2000-3000 pmolIl, flushing, tachycardia and a reduction in diastolic blood pressure. Peptide histidine isoleucine caused a significant inhibition of net absorption of water, sodium, potassium and bicarbonate and induced a net secretion of chloride, these changes being completely reversed during the post-peptide histidine isoleucine period. These findings suggest that endogenous peptide histidine isoleucine may participate in the neurohumoral regulation of water and electrolyte transport in the human jejunum. http://gut.bmj.com/ Peptide histidine isoleucine, isolated originally from INTESTINAL PERFUSION mammalian small intestine, is a 27-amino acid After an eight hour fast, each subject swallowed a peptide having close amino acid sequence homology double lumen intestinal perfusion tube, incorpo- to vasoactive intestinal peptide and secretin. -

Al Exposure Increases Proline Levels by Different Pathways in An
www.nature.com/scientificreports OPEN Al exposure increases proline levels by diferent pathways in an Al‑sensitive and an Al‑tolerant rye genotype Alexandra de Sousa1,2, Hamada AbdElgawad2,4, Fernanda Fidalgo1, Jorge Teixeira1, Manuela Matos3, Badreldin A. Hamed4, Samy Selim5, Wael N. Hozzein6, Gerrit T. S. Beemster2 & Han Asard2* Aluminium (Al) toxicity limits crop productivity, particularly at low soil pH. Proline (Pro) plays a role in protecting plants against various abiotic stresses. Using the relatively Al‑tolerant cereal rye (Secale cereale L.), we evaluated Pro metabolism in roots and shoots of two genotypes difering in Al tolerance, var. RioDeva (sensitive) and var. Beira (tolerant). Most enzyme activities and metabolites of Pro biosynthesis were analysed. Al induced increases in Pro levels in each genotype, but the mechanisms were diferent and were also diferent between roots and shoots. The Al‑tolerant genotype accumulated highest Pro levels and this stronger increase was ascribed to simultaneous activation of the ornithine (Orn)‑biosynthetic pathway and decrease in Pro oxidation. The Orn pathway was particularly enhanced in roots. Nitrate reductase (NR) activity, N levels, and N/C ratios demonstrate that N‑metabolism is less inhibited in the Al‑tolerant line. The correlation between Pro changes and diferences in Al‑sensitivity between these two genotypes, supports a role for Pro in Al tolerance. Our results suggest that diferential responses in Pro biosynthesis may be linked to N‑availability. Understanding the role of Pro in diferences between genotypes in stress responses, could be valuable in plant selection and breeding for Al resistance. Proline (Pro) is involved in a wide range of plant physiological and developmental processes1. -
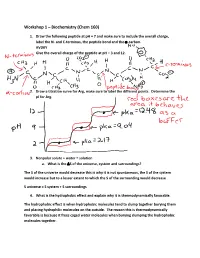
Workshop 1 – Biochemistry (Chem 160)
Workshop 1 – Biochemistry (Chem 160) 1. Draw the following peptide at pH = 7 and make sure to include the overall charge, label the N- and C-terminus, the peptide bond and the -carbon. AVDKY Give the overall charge of the peptide at pH = 3 and 12. 2. Draw a titration curve for Arg, make sure to label the different points. Determine the pI for Arg. 3. Nonpolar solute + water = solution a. What is the S of the universe, system and surroundings? The S of the universe would decrease this is why it is not spontaneous, the S of the system would increase but to a lesser extent to which the S of the surrounding would decrease S universe = S system + S surroundings 4. What is the hydrophobic effect and explain why it is thermodynamically favorable. The hydrophobic effect is when hydrophobic molecules tend to clump together burying them and placing hydrophilic molecules on the outside. The reason this is thermodynamically favorable is because it frees caged water molecules when burying clumping the hydrophobic molecules together. 5. Urea dissolves very readily in water, but the solution becomes very cold as the urea dissolves. How is this possible? Urea dissolves in water because when dissolving there is a net increase in entropy of the universe. The heat exchange, getting colder only reflects the enthalpy (H) component of the total energy change. The entropy change is high enough to offset the enthalpy component and to add up to an overall -G 6. A mutation that changes an alanine residue in the interior of a protein to valine is found to lead to a loss of activity. -

Thermal Decomposition of the Amino Acids Glycine, Cysteine, Aspartic Acid, Asparagine, Glutamic Acid, Glutamine, Arginine and Histidine
bioRxiv preprint doi: https://doi.org/10.1101/119123; this version posted March 22, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Thermal decomposition of the amino acids glycine, cysteine, aspartic acid, asparagine, glutamic acid, glutamine, arginine and histidine Ingrid M. Weiss*, Christina Muth, Robert Drumm & Helmut O.K. Kirchner INM-Leibniz Institute for New Materials, Campus D2 2, D-66123 Saarbruecken Germany *Present address: Universität Stuttgart, Institut für Biomaterialien und biomolekulare Systeme, Pfaffenwaldring 57, D-70569 Stuttgart, Germany Abstract Calorimetry, thermogravimetry and mass spectrometry were used to follow the thermal decomposition of the eight amino acids G, C, D, N, E, Q, R and H between 185°C and 280°C. Endothermic heats of decomposition between 72 and 151 kJ/mol are needed to form 12 to 70 % volatile products. This process is neither melting nor sublimation. With exception of cysteine they emit mainly H2O, some NH3 and no CO2. Cysteine produces CO2 and little else. The reactions are described by polynomials, AA → a (NH3) + b (H2O) + c (CO2) + d (H2S) + e (residue), with integer or half integer coefficients. The solid monomolecular residues are rich in peptide bonds. 1. Motivation Amino acids might have been synthesized under prebiological conditions on earth or deposited on earth from interstellar space, where they have been found [Follmann and Brownson, 2009]. Robustness of amino acids against extreme conditions is required for early occurrence, but little is known about their nonbiological thermal destruction. There is hope that one might learn something about the molecules needed in synthesis from the products found in decomposition. -

Effects of Single Amino Acid Deficiency on Mrna Translation Are Markedly
www.nature.com/scientificreports OPEN Efects of single amino acid defciency on mRNA translation are markedly diferent for methionine Received: 12 December 2016 Accepted: 4 May 2018 versus leucine Published: xx xx xxxx Kevin M. Mazor, Leiming Dong, Yuanhui Mao, Robert V. Swanda, Shu-Bing Qian & Martha H. Stipanuk Although amino acids are known regulators of translation, the unique contributions of specifc amino acids are not well understood. We compared efects of culturing HEK293T cells in medium lacking either leucine, methionine, histidine, or arginine on eIF2 and 4EBP1 phosphorylation and measures of mRNA translation. Methionine starvation caused the most drastic decrease in translation as assessed by polysome formation, ribosome profling, and a measure of protein synthesis (puromycin-labeled polypeptides) but had no signifcant efect on eIF2 phosphorylation, 4EBP1 hyperphosphorylation or 4EBP1 binding to eIF4E. Leucine starvation suppressed polysome formation and was the only tested condition that caused a signifcant decrease in 4EBP1 phosphorylation or increase in 4EBP1 binding to eIF4E, but efects of leucine starvation were not replicated by overexpressing nonphosphorylatable 4EBP1. This suggests the binding of 4EBP1 to eIF4E may not by itself explain the suppression of mRNA translation under conditions of leucine starvation. Ribosome profling suggested that leucine deprivation may primarily inhibit ribosome loading, whereas methionine deprivation may primarily impair start site recognition. These data underscore our lack of a full -

Amino Acid Recognition by Aminoacyl-Trna Synthetases
www.nature.com/scientificreports OPEN The structural basis of the genetic code: amino acid recognition by aminoacyl‑tRNA synthetases Florian Kaiser1,2,4*, Sarah Krautwurst3,4, Sebastian Salentin1, V. Joachim Haupt1,2, Christoph Leberecht3, Sebastian Bittrich3, Dirk Labudde3 & Michael Schroeder1 Storage and directed transfer of information is the key requirement for the development of life. Yet any information stored on our genes is useless without its correct interpretation. The genetic code defnes the rule set to decode this information. Aminoacyl-tRNA synthetases are at the heart of this process. We extensively characterize how these enzymes distinguish all natural amino acids based on the computational analysis of crystallographic structure data. The results of this meta-analysis show that the correct read-out of genetic information is a delicate interplay between the composition of the binding site, non-covalent interactions, error correction mechanisms, and steric efects. One of the most profound open questions in biology is how the genetic code was established. While proteins are encoded by nucleic acid blueprints, decoding this information in turn requires proteins. Te emergence of this self-referencing system poses a chicken-or-egg dilemma and its origin is still heavily debated 1,2. Aminoacyl-tRNA synthetases (aaRSs) implement the correct assignment of amino acids to their codons and are thus inherently connected to the emergence of genetic coding. Tese enzymes link tRNA molecules with their amino acid cargo and are consequently vital for protein biosynthesis. Beside the correct recognition of tRNA features3, highly specifc non-covalent interactions in the binding sites of aaRSs are required to correctly detect the designated amino acid4–7 and to prevent errors in biosynthesis5,8. -

Relative Reaction Rates of the Amino Acids Cysteine, Methionine, and Histidine with Analogs of the Anti-Cancer Drug Cisplatin Cynthia A
Western Kentucky University TopSCHOLAR® Honors College Capstone Experience/Thesis Honors College at WKU Projects 5-11-2015 Relative Reaction Rates of the Amino Acids Cysteine, Methionine, and Histidine with Analogs of the Anti-Cancer Drug Cisplatin Cynthia A. Tope Western Kentucky University, [email protected] Follow this and additional works at: http://digitalcommons.wku.edu/stu_hon_theses Part of the Medicinal-Pharmaceutical Chemistry Commons Recommended Citation Tope, Cynthia A., "Relative Reaction Rates of the Amino Acids Cysteine, Methionine, and Histidine with Analogs of the Anti-Cancer Drug Cisplatin" (2015). Honors College Capstone Experience/Thesis Projects. Paper 571. http://digitalcommons.wku.edu/stu_hon_theses/571 This Thesis is brought to you for free and open access by TopSCHOLAR®. It has been accepted for inclusion in Honors College Capstone Experience/ Thesis Projects by an authorized administrator of TopSCHOLAR®. For more information, please contact [email protected]. RELATIVE REACTION RATES OF THE AMINO ACIDS CYSTEINE, METHIONINE, AND HISTIDINE WITH ANALOGS OF THE ANTI-CANCER DRUG CISPLATIN A Capstone Experience/Thesis Project Presented in Partial Fulfillment of the Requirements for the Degree Bachelor of Science with Honors College Graduate Distinction at Western Kentucky University By: Cynthia A. Tope ***** Western Kentucky University 2015 CE/T Committee: Approved by: Professor Kevin Williams, Advisor _________________________ Professor Darwin Dahl Advisor Professor Lee Ann Smith Department of Chemistry Copyright: Cynthia A. Tope 2015 ABSTRACT We are studying the reaction of analogs of the anticancer drug cisplatin with amino acids that differ in size and shape. The reaction of cisplatin with proteins likely precedes reaction with DNA in the body, forming a variety of products that may be toxic to the human body. -

Conformational Properties of Constrained Proline Analogues and Their Application in Nanobiology”
UNIVERSITAT POLITÈCNICA DE CATALUNYA DEPARTAMENT D’ENGINYERIA QUÍMICA “CONFORMATIONAL PROPERTIES OF CONSTRAINED PROLINE ANALOGUES AND THEIR APPLICATION IN NANOBIOLOGY” Alejandra Flores Ortega Supervisors: Dr. Carlos Alemán Llansó and Dr. Jordi Casanovas Salas. th Barcelona, 27 January 2009 “Chance is a word void of sense; nothing can exist without a cause”. François-Marie Arouet, Voltaire “Imagination will often carry us to worlds that never were. But without it, we go nowhere”. Carl Sagan iii ACKNOWLEDGEMENTS I would like to acknowledge to Dr. Carlos Aleman and Dr. Jordi Cassanovas Salas for an interesting research theme, and scientific support. I gratefully acknowledge to Dr. David Zanuy for interesting suggestions and strong discussions, without their support this would be an unfulfilled task. Also I, would like to address my thanks to all my colleagues in my group and department, specially Elaine Armelin for assiting me in many different ways. I thank not only my friends, but also colleagues for helping me to overcome the stressful time, without whom it would have been difficult to cope up. I wish to express my gratefulness to my parents, specially to my mother, María Esther, for all his care, and support. Also I will like to thanks to my friends and specially Jesus, Merches, Laura y Arturo. My PhD thesis have been finished for all this support. I am greatly indepted to Dr. Ruth Nussinov at NCI, Dr. Carlos Cativiela at the University of Zaragoza and Ana I. Jiménez at the “Instituto de Ciencias de Materiales de Aragon” for a collaborative effort. I wish to thank all my colleague in the “Chimie et Biochimie Théoriques, Faculté des Sciences et Techniques” in Nancy France, I will be grateful to have worked with : Pr. -

Asparagine-Proline Sequence Within Membrane-Spanning Segment of SREBP Triggers Intramembrane Cleavage by Site-2 Protease
Asparagine-proline sequence within membrane-spanning segment of SREBP triggers intramembrane cleavage by Site-2 protease Jin Ye*†, Utpal P. Dave´ *†, Nick V. Grishin‡, Joseph L. Goldstein*§, and Michael S. Brown*§ Departments of *Molecular Genetics and ‡Biochemistry, University of Texas Southwestern Medical Center, 5323 Harry Hines Boulevard, Dallas, TX 75390-9046 Contributed by Joseph L. Goldstein, March 16, 2000 The NH2-terminal domains of membrane-bound sterol regulatory nus. It translocates to the nucleus, where it activates more than element-binding proteins (SREBPs) are released into the cytosol by 20 genes encoding enzymes of cholesterol and fatty acid synthesis regulated intramembrane proteolysis, after which they enter the as well as the low density lipoprotein receptor (6, 7). When nucleus to activate genes encoding lipid biosynthetic enzymes. sterols build up in cells, the SREBP͞SCAP complex fails to exit Intramembrane proteolysis is catalyzed by Site-2 protease (S2P), a the ER, and it never reaches S1P (8, 9). As a result, the hydrophobic zinc metalloprotease that cleaves SREBPs at a mem- NH2-terminal domains of the SREBPs are no longer released brane-embedded leucine-cysteine bond. In the current study, we into the nucleus, and transcription of the target genes declines. use domain-swapping methods to localize the residues within This mechanism allows cholesterol to inhibit its own synthesis the SREBP-2 membrane-spanning segment that are required for and uptake, thereby preventing cholesterol overaccumulation in cleavage by S2P. The studies reveal a requirement for an asparag- cells. ine-proline sequence in the middle third of the transmembrane The human gene encoding S2P was cloned by complementa- segment. -

Electrochemical Studies of Dl-Leucine, L-Proline and L
ELECTROCHEMICAL STUDIES OF DL -LEUCINE, 60 L-PROLINE AND L-TRYPTOPHAN AND THEIR INTERACTION WITH COPPER AND IRON 30 c b A) M. A. Jabbar, R. J. Mannan, S. Salauddin and B. µ a Rashid 0 Department of Chemistry, University of Dhaka, ( Current Dhaka-1000, Bangladesh -30 Introduction -60 In vitro study of the charge transfer reactions coupled -800 -400 0 400 800 with chemical reactions can give important indication of Potential vs. Ag/AgCl (mV) about actual biological processes occurring in human Fig.1. Comparison of the cyclic voltammogram of system. Understanding of such charge-transfer 5.0mM (a) DL -Leucine, (b) Cu-DL -Leucine ion and mechanism will help to determine the effectiveness of (c) [Fe-DL -Leucine] in 0.1M KCl solution at a Pt- nutrition, metabolism and treatment of various biological button electrode. Scan rates 50 mV/s. disorders. In the previous research, the redox behaviour of 40 various amino acids and biochemically important compounds and their charge transfer reaction and their b interaction of metal ions were studied [1,2]. In the present 20 ) a c research, the redox behavior and the charge transfer µΑ kinetics of DL -Leucine, L-Proline and L-Tryptophan in 0 the presence and absence of copper and iron will be investigated. Current ( -20 Experimental A computerized electrochemistry system developed by -40 -800 -400 0 400 800 Advanced Analytics, Virginia, USA, (Model-2040) Potential vs. Ag/AgCl (mV) consisting of three electrodes micro-cell with a saturated Ag/AgCl reference, a Pt-wire auxiliary and a pretreated Fig.2 . Comparison of the cyclic voltammogram of Pt-button working electrode is employed to investigate 5.0mM (a) L-Proline, (b) Cu-L-Proline and (c) Fe-L- Proline in 0.1M KCl solution at a Pt-button different amino acids and metal-amino acid systems. -

Convenient Preparation of Poly(L-Histidine) by the Direct Polymerization of L-Histidine Or Nim Benzyl-L-Histidine with Diphenylphosphoryl Azide
Polymer Journal, Vol. 13, No. 12, pp 1151-1154 (1981) SHORT COMMUNICATION Convenient Preparation of Poly(L-histidine) by the Direct Polymerization of L-Histidine or Nim_Benzyl-L-histidine with Diphenylphosphoryl Azide Takumi NARUSE, Bun-ichiro NAKAJIMA, Akihiro TSUTSUMI, and Norio NISHI Department of Polymer Science, Faculty of Science, Hokkaido University, Nishi 8-chome, Kita 10-jo, Kita-ku, Sapporo 060, Japan. (Received August 14, 1981) KEY WORDS Polymerization I Diphenylphosphoryl Azide (DPPA) I L- Histidine I N'm-Benzyl-L-histidine I Poly(L-histidine) I Poly(N'm-benzyl-L histidine) I IR Spectrum I 13C NMR Spectrum I Intrinsic Viscosity I Poly(L-histidine) is a very interesting poly(a histidine) by the polymerization of L-histidine with amino acid) as a synthetic functional polymer or as iodine-phosphonic acid esters was unsuccessful. The a model for the functional biopolymer such as simplification of the synthetic route for poly(L enzymes. It is known, however, that poly(L histidine) is still necessary. histidine) can not be prepared by the Fuchs Diphenylphosphoryl azide (DPPA) has been used Farthing method 1 which is very popular for prepar as a convenient reagent for racemization-free pep ing poly(a-amino acid)s. A synthetic route with tide synthesis, since it was synthesized by Shioiri et several steps involving the synthesis of NCA (a a!. in 1972.6 Recently, we reported that this reagent amino acid N-carboxyanhydride) by the rather can also be used successfully for the polymerization classical Leuchs method/ have been used for the of amino acids or peptides.