Target-Genes Reveal Species and Genotypic Specificity of Anthocyanin Pigmentation in Citrus and Related Genera
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Fruits; Fresh Vegetables and Fresh Limes” (Opp
Trademark Trial and Appeal Board Electronic Filing System. http://estta.uspto.gov ESTTA Tracking number: ESTTA881622 Filing date: 03/07/2018 IN THE UNITED STATES PATENT AND TRADEMARK OFFICE BEFORE THE TRADEMARK TRIAL AND APPEAL BOARD Proceeding 91238258 Party Plaintiff Wonderful Citrus LLC Correspondence DARYA P LAUFER ESQ Address ROLL LAW GROUP PC 11444 WEST OLYMPIC BLVD LOS ANGELES, CA 90064 UNITED STATES Email: [email protected], [email protected] Submission Other Motions/Papers Filer's Name Michael M. Vasseghi Filer's email [email protected], [email protected] Signature / Michael M. Vasseghi / Date 03/07/2018 Attachments Opposition with Exhibits-reduced size.pdf(1950576 bytes ) IN THE UNITED STATES PATENT AND TRADEMARK OFFICE TRADEMARK TRIAL AND APPEAL BOARD Wonderful Citrus LLC, Opposition No. 91238258 Opposer, Application Serial No. 87/472272 v. APB, Inc. dba Vision Produce Company, Applicant. OPPOSER WONDERFUL CITRUS LLC’S OPPOSITION TO APPLICANT’S MOTION FOR JUDGMENT ON THE PLEADINGS I. INTRODUCTION Applicant moves for judgment on the pleadings (“Motion”), arguing that “there is no genuine issue as to Opposer’s lack of prior rights in a trademark that could be confusingly similar to Applicant’s Mark.” (Motion pg. 3.)1 Applicant’s Motion is not well taken. It acknowledges that Opposer has alleged exactly what it takes issue with – that Opposer has prior rights in a trademark that could be confusingly similar to Applicant’s Mark. Despite this, Applicant seeks to take issue with those allegations, implicitly contending that Opposer will be unable to prove what it has alleged. (Motion pg. 2.) This is not a proper basis for judgment on the pleadings, which must accept as true all allegations asserted in the Opposition. -

UNIVERSITY of CALIFORNIA RIVERSIDE Cross-Compatibility, Graft-Compatibility, and Phylogenetic Relationships in the Aurantioi
UNIVERSITY OF CALIFORNIA RIVERSIDE Cross-Compatibility, Graft-Compatibility, and Phylogenetic Relationships in the Aurantioideae: New Data From the Balsamocitrinae A Thesis submitted in partial satisfaction of the requirements for the degree of Master of Science in Plant Biology by Toni J Siebert Wooldridge December 2016 Thesis committee: Dr. Norman C. Ellstrand, Chairperson Dr. Timothy J. Close Dr. Robert R. Krueger The Thesis of Toni J Siebert Wooldridge is approved: Committee Chairperson University of California, Riverside ACKNOWLEDGEMENTS I am indebted to many people who have been an integral part of my research and supportive throughout my graduate studies: A huge thank you to Dr. Norman Ellstrand as my major professor and graduate advisor, and to my supervisor, Dr. Tracy Kahn, who helped influence my decision to go back to graduate school while allowing me to continue my full-time employment with the UC Riverside Citrus Variety Collection. Norm and Tracy, my UCR parents, provided such amazing enthusiasm, guidance and friendship while I was working, going to school and caring for my growing family. Their support was critical and I could not have done this without them. My committee members, Dr. Timothy Close and Dr. Robert Krueger for their valuable advice, feedback and suggestions. Robert Krueger for mentoring me over the past twelve years. He was the first person I met at UCR and his willingness to help expand my knowledge base on Citrus varieties has been a generous gift. He is also an amazing friend. Tim Williams for teaching me everything I know about breeding Citrus and without whom I'd have never discovered my love for the art. -

Known Host Plants of Huanglongbing (HLB) and Asian Citrus Psyllid
Known Host Plants of Huanglongbing (HLB) and Asian Citrus Psyllid Diaphorina Liberibacter citri Plant Name asiaticus Citrus Huanglongbing Psyllid Aegle marmelos (L.) Corr. Serr.: bael, Bengal quince, golden apple, bela, milva X Aeglopsis chevalieri Swingle: Chevalier’s aeglopsis X X Afraegle gabonensis (Swingle) Engl.: Gabon powder-flask X Afraegle paniculata (Schum.) Engl.: Nigerian powder- flask X Atalantia missionis (Wall. ex Wight) Oliv.: see Pamburus missionis X X Atalantia monophylla (L.) Corr.: Indian atalantia X Balsamocitrus dawei Stapf: Uganda powder- flask X X Burkillanthus malaccensis (Ridl.) Swingle: Malay ghost-lime X Calodendrum capense Thunb.: Cape chestnut X × Citroncirus webberi J. Ingram & H. E. Moore: citrange X Citropsis gilletiana Swingle & M. Kellerman: Gillet’s cherry-orange X Citropsis schweinfurthii (Engl.) Swingle & Kellerm.: African cherry- orange X Citrus amblycarpa (Hassk.) Ochse: djerook leemo, djeruk-limau X Citrus aurantiifolia (Christm.) Swingle: lime, Key lime, Persian lime, lima, limón agrio, limón ceutí, lima mejicana, limero X X Citrus aurantium L.: sour orange, Seville orange, bigarde, marmalade orange, naranja agria, naranja amarga X Citrus depressa Hayata: shiikuwasha, shekwasha, sequasse X Citrus grandis (L.) Osbeck: see Citrus maxima X Citrus hassaku hort. ex Tanaka: hassaku orange X Citrus hystrix DC.: Mauritius papeda, Kaffir lime X X Citrus ichangensis Swingle: Ichang papeda X Citrus jambhiri Lushington: rough lemon, jambhiri-orange, limón rugoso, rugoso X X Citrus junos Sieb. ex Tanaka: xiang -

Effect of Temperature on Germination of Citrus Macroptera, Citrus Latipes and Citrus Indica Seeds *Anamika Upadhaya, Shiva S
ISSN. 0972 - 8406 The NEHU Journal Vol. XVII, No. 1 (January - June) and No. 2 (July - December) 2019, pp. 12-20 Effect of temperature on germination of Citrus macroptera, Citrus latipes and Citrus indica seeds *Anamika Upadhaya, Shiva S. Chaturvedi, Brajesh K. Tiwari and Dibyendu Paul Department of Environmental Studies, North Eastern Hill University Umshing, Meghalaya, India – 793022 *Corresponding author : [email protected] Abstract Seeds are an important means of propagation of Citrus species. Seeds of three wild Citrus namely; Citrus macroptera Montrouz., Citrus latipes (Swingle) Tanaka and Citrus indica Tanaka were germinated at 20°C, 25°C, 30°C and 35°C temperature to observe the effect of temperature on germination. Mean germination time and percentage seed germinated were recorded and used to determine optimum temperature for germination. Viability of seeds determined using chemical and germination tests yielded similar results. Optimum temperature for germination was found to be 28°C for C. macroptera and C. latipes and 26°C for C. indica. Keywords: Germination, wild, C. macroptera, C. latipes, C. indica, Meghalaya Introduction Citrus has been domesticated since ancient times, and where ‘natural’ populations are located, it is often difficult to determine whether they represent wild ancestors or are derived from naturalized forms of introduced varieties. Though relatively rare in wild, Citrus are mostly found as scattered trees in primary forests in remote areas rather than as pure stands. In India, a vast reservoir of Citrus diversity exists both in wild and in cultivated forms. North-eastern India is considered as natural home of many Citrus species with wide occurrence of indigenous species like C. -

Chemical Variability of Peel and Leaf Essential Oils in the Citrus Subgenus Papeda (Swingle) and Relatives
Chemical variability of peel and leaf essential oils in the Citrus subgenus Papeda (Swingle) and relatives Clémentine Baccati, Marc Gibernau, Mathieu Paoli, Patrick Ollitrault, Félix Tomi, François Luro To cite this version: Clémentine Baccati, Marc Gibernau, Mathieu Paoli, Patrick Ollitrault, Félix Tomi, et al.. Chemical variability of peel and leaf essential oils in the Citrus subgenus Papeda (Swingle) and relatives. Plants, MDPI, 2021, 10 (6), pp.1117. 10.3390/plants10061117. hal-03262123 HAL Id: hal-03262123 https://hal.archives-ouvertes.fr/hal-03262123 Submitted on 16 Jun 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License Chemical variability of peel and leaf essential oils in the Citrus subgenus Papeda (Swingle) and relatives Clémentine Baccati 1, Marc Gibernau 1, Mathieu Paoli 1, Patrick Ollitrault 2,3, Félix Tomi 1, * and François Luro 2 1 Université de Corse-CNRS, UMR 6134 SPE, Route des Sanguinaires, 20000 Ajaccio, France; [email protected] (C.B.) ; [email protected] (M.G.) ; [email protected] (M.P.) ; [email protected] (F.T.) 2 UMR AGAP Institut, Univ Montpellier, CIRAD, INRAE, Institut Agro – 20230, San Giuliano, France 3 CIRAD, UMR AGAP, F-20230 San Giuliano, France * Correspondence: [email protected]; tel.:+33-495-52-4122. -
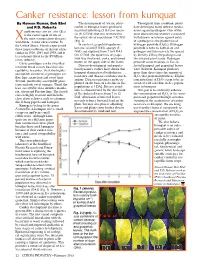
Canker Resistance: Lesson from Kumquat by Naveen Kumar, Bob Ebel the Development of Asiatic Citrus Throughout Their Evolution, Plants and P.D
Canker resistance: lesson from kumquat By Naveen Kumar, Bob Ebel The development of Asiatic citrus Throughout their evolution, plants and P.D. Roberts canker in kumquat leaves produced have developed many defense mecha- anthomonas citri pv. citri (Xcc) localized yellowing (5 DAI) or necro- nisms against pathogens. One of the is the causal agent of one of sis (9-12 DAI) that was restricted to most characteristic features associated the most serious citrus diseases the actual site of inoculation 7-12 DAI with disease resistance against entry X (Fig. 2). of a pathogen is the production of worldwide, Asiatic citrus canker. In the United States, Florida experienced In contrast, grapefruit epidermis hydrogen peroxide (H2O2). Hydrogen three major outbreaks of Asiatic citrus became raised (5 DAI), spongy (5 peroxide is toxic to both plant and canker in 1910, 1984 and 1995, and it DAI) and ruptured from 7 to 8 DAI. pathogen and thus restricts the spread is a constant threat to the $9 billion On 12 DAI, the epidermis of grape- by directly killing the pathogen and citrus industry. fruit was thickened, corky, and turned the infected plant tissue. Hydrogen Citrus genotypes can be classified brown on the upper side of the leaves. peroxide concentrations in Xcc-in- into four broad classes based on sus- Disease development and popula- fected kumquat and grapefruit leaves ceptibility to canker. First, the highly- tion dynamics studies have shown that were different. Kumquat produces susceptible commercial genotypes are kumquat demonstrated both disease more than three times the amount of Key lime, grapefruit and sweet lime. -

Freeze Response of Citrus and Citrus- Speeds (Nisbitt Et Al., 2000)
HORTSCIENCE 49(8):1010–1016. 2014. and tree and grove size (Bourgeois et al., 1990; Ebel et al., 2005). Protection using microsprinklers is compromised by high wind Freeze Response of Citrus and Citrus- speeds (Nisbitt et al., 2000). Developing more cold-tolerant citrus varieties through breeding related Genotypes in a Florida Field and selection has long been considered the most effective long-term solution (Grosser Planting et al., 2000; Yelenosky, 1985). Citrus and Citrus relatives are members Sharon Inch, Ed Stover1, and Randall Driggers of the family Rutaceae. The subtribe Citrinae U.S. Horticultural Research Laboratory, U.S. Department of Agriculture, is composed of Citrus (mandarins, oranges, Agricultural Research Service, 2001 South Rock Road, Fort Pierce, FL pummelos, grapefruits, papedas, limes, lem- ons, citrons, and sour oranges); Poncirus 34945 (deciduous trifoliate oranges); Fortunella Richard F. Lee (kumquats); Microcitrus and Eremocitrus (both Australian natives); and Clymenia National Clonal Germplasm Repository for Citrus and Dates, U.S. (Penjor et al., 2013). There is considerable Department of Agriculture, Agricultural Research Service, 1060 Martin morphological and ecological variation within Luther King Boulevard, Riverside, CA 92521 this group. With Citrus, cold-hardiness ranges from cold-tolerant to cold-sensitive (Soost and Additional index words. Aurantioideae, citrus breeding, cold-sensitive, defoliation, dieback, Roose, 1996). Poncirus and Fortunella are frost damage, Rutaceae, Toddalioideae considered the most cold-tolerant genera that Abstract. A test population consisting of progenies of 92 seed-source genotypes (hereafter are cross-compatible with Citrus. Poncirus called ‘‘parent genotypes’’) of Citrus and Citrus relatives in the field in east–central trifoliata reportedly can withstand tempera- Florida was assessed after natural freeze events in the winters of 2010 and 2011. -

PRUNING GUIDELINES Tools: Loppers, Saw, Clippers 10% Bleach and Water Solution
Varieties - choose one that you will want to eat often, as you will have them much of the year (Four Winds Citrus Variety Chart link in your Resources list) - certain citrus mature earlier than others (see early ripening handout) - Unique varieties: - Blood orange: red flesh is antioxidant rich. Often sweeter than other oranges - Yuzu: very little, but very flavorful juice used by chefs. Believed to be a cross between a sour mandarin and ichang papeda - Keiffir lime: regular lime with bumpy skin. Attractive tree with segmented leaves that are extremely fragrant and prized by chefs. - Buddhas hand: not much juice, but very fragrant pith and rind. Odd shaped and often used as an ornamental - Australian finger lime: oblong green lime with many small lime-flavored orbs inside. Called "citrus caviar" Standard varieties of citrus trees often grow to a height of 20 to 30 feet and the canopy -- or width of a tree -- can spread to 18 to 30 feet depending on the variety. Dwarf citrus trees are significantly shorter and narrower, which provides greater flexibility in planting location. Most varieties top out at 8 feet in height with a proportionally smaller canopy. Despite the differences in height and width, regular, semi-dwarf and dwarf citrus varieties produce the same size fruit. -First off DO NOT PRUNE! Those damaged leaves can actually provide protection for the plant until the air warms up. The plant needs to rally and recover. Pruning might just put it over the edge. Sometimes the plants must remain with that ‘raggedy’ appearance until as late as June and in some cases a full year. -

Citrus Industry Biosecurity Plan 2015
Industry Biosecurity Plan for the Citrus Industry Version 3.0 July 2015 PLANT HEALTH AUSTRALIA | Citrus Industry Biosecurity Plan 2015 Location: Level 1 1 Phipps Close DEAKIN ACT 2600 Phone: +61 2 6215 7700 Fax: +61 2 6260 4321 E-mail: [email protected] Visit our web site: www.planthealthaustralia.com.au An electronic copy of this plan is available through the email address listed above. © Plant Health Australia Limited 2004 Copyright in this publication is owned by Plant Health Australia Limited, except when content has been provided by other contributors, in which case copyright may be owned by another person. With the exception of any material protected by a trade mark, this publication is licensed under a Creative Commons Attribution-No Derivs 3.0 Australia licence. Any use of this publication, other than as authorised under this licence or copyright law, is prohibited. http://creativecommons.org/licenses/by-nd/3.0/ - This details the relevant licence conditions, including the full legal code. This licence allows for redistribution, commercial and non-commercial, as long as it is passed along unchanged and in whole, with credit to Plant Health Australia (as below). In referencing this document, the preferred citation is: Plant Health Australia Ltd (2004) Industry Biosecurity Plan for the Citrus Industry (Version 3.0 – July 2015). Plant Health Australia, Canberra, ACT. Disclaimer: The material contained in this publication is produced for general information only. It is not intended as professional advice on any particular matter. No person should act or fail to act on the basis of any material contained in this publication without first obtaining specific and independent professional advice. -

Joimalofagmoiltdraiesea
JOIMALOFAGMOILTDRAIESEARCH VOL. XIX WASHINGTON, D. C, JULY 15, 1920 No. 8 RELATIVE SUSCEPTIBILITY TO CITRUS-CANKER OF DIFFERENT SPECIES AND HYBRIDS OF THE GENUS CITRUS, INCLUDING THE WILD RELATIVES » By GEORGE I*. PELTIER, Plant Pathologist, Alabama Agricultural Experiment Station, and Agent, Bureau of Plant Industry, United States Department of Agriculture, and WILLIAM J. FREDERICH, Assistant Pathologist, Bureau of Plant Industry, United States Department of Agriculture 2 INTRODUCTION In a preliminary report (6)3 the senior author briefly described the results obtained under greenhouse conditions for a period of six months on the susceptibility and resistance to citrus-canker of a number of plants including some of the wild relatives, Citrus fruits, and hybrids of the genus Citrus. Since that time the plants reported on have been under close observation; a third experiment has been started, and many inoculations have been made in the isolation field in southern Alabama during the summers of 1917, 1918, and 1919. Many more plants have been successfully inoculated; others have proved to be extremely sus- ceptible; while some of those tested still show considerable resistance. The results obtained up to November 1, 1919, are described in tjhis report. EXPERIMENTAL METHODS In the greenhouse, the methods used and the conditions governing the inoculations described in the preliminary report were closely fol- lowed. The same strain of the organism was used and was applied in the 1 Published with the approval of the Director of the Alabama Agricultural Experiment Station. The paper is based upon cooperative investigations between the Office of Crop Physiology and Breeding Investi- gations, Bureau of Plant Industry, United States Department of Agriculture, and the Department of Plant Pathology, Alabama Agricultural Experiment Station. -
Holdings of the University of California Citrus Variety Collection 41
Holdings of the University of California Citrus Variety Collection Category Other identifiers CRC VI PI numbera Accession name or descriptionb numberc numberd Sourcee Datef 1. Citron and hybrid 0138-A Indian citron (ops) 539413 India 1912 0138-B Indian citron (ops) 539414 India 1912 0294 Ponderosa “lemon” (probable Citron ´ lemon hybrid) 409 539491 Fawcett’s #127, Florida collection 1914 0648 Orange-citron-hybrid 539238 Mr. Flippen, between Fullerton and Placentia CA 1915 0661 Indian sour citron (ops) (Zamburi) 31981 USDA, Chico Garden 1915 1795 Corsican citron 539415 W.T. Swingle, USDA 1924 2456 Citron or citron hybrid 539416 From CPB 1930 (Came in as Djerok which is Dutch word for “citrus” 2847 Yemen citron 105957 Bureau of Plant Introduction 3055 Bengal citron (ops) (citron hybrid?) 539417 Ed Pollock, NSW, Australia 1954 3174 Unnamed citron 230626 H. Chapot, Rabat, Morocco 1955 3190 Dabbe (ops) 539418 H. Chapot, Rabat, Morocco 1959 3241 Citrus megaloxycarpa (ops) (Bor-tenga) (hybrid) 539446 Fruit Research Station, Burnihat Assam, India 1957 3487 Kulu “lemon” (ops) 539207 A.G. Norman, Botanical Garden, Ann Arbor MI 1963 3518 Citron of Commerce (ops) 539419 John Carpenter, USDCS, Indio CA 1966 3519 Citron of Commerce (ops) 539420 John Carpenter, USDCS, Indio CA 1966 3520 Corsican citron (ops) 539421 John Carpenter, USDCS, Indio CA 1966 3521 Corsican citron (ops) 539422 John Carpenter, USDCS, Indio CA 1966 3522 Diamante citron (ops) 539423 John Carpenter, USDCS, Indio CA 1966 3523 Diamante citron (ops) 539424 John Carpenter, USDCS, Indio -

Bergamot Oil: Botany, Production, Pharmacology
Entry Bergamot Oil: Botany, Production, Pharmacology Marco Valussi 1,* , Davide Donelli 2 , Fabio Firenzuoli 3 and Michele Antonelli 2 1 Herbal and Traditional Medicine Practitioners Association (EHTPA), Norwich NR3 1HG, UK 2 AUSL-IRCCS Reggio Emilia, 42122 Reggio Emilia RE, Italy; [email protected] (D.D.); [email protected] (M.A.) 3 CERFIT, Careggi University Hospital, 50139 Firenze FI, Italy; fabio.firenzuoli@unifi.it * Correspondence: [email protected] Definition: Bergamot essential oil (BEO) is the result of the mechanical manipulation (cold pressing) of the exocarp (flavedo) of the hesperidium of Citrus limon (L.) Osbeck Bergamot Group (synonym Citrus × bergamia Risso & Poit.), resulting in the bursting of the oil cavities embedded in the flavedo and the release of their contents. It is chemically dominated by monoterpene hydrocarbons (i.e., limonene), but with significant percentages of oxygenated monoterpenes (i.e., linalyl acetate) and of non-volatile oxygen heterocyclic compounds (i.e., bergapten). Keywords: bergamot; citrus; essential oil; production; review 1. Introduction The taxonomy, and consequently the nomenclature, of the genus Citrus, is particularly complicated and has been rapidly changing in recent years (see Table1). For over 400 years, the centre of origin and biodiversity, along with the evolution and phylogeny of the Citrus species, have all been puzzling problems for botanists and the confusing and changing Citation: Valussi, M.; Donelli, D.; nomenclature of this taxon over the years can reflect intrinsic reproductive features of Firenzuoli, F.; Antonelli, M. Bergamot Oil: Botany, Production, the species included in this genus, the cultural and geographical issues, and the rapidly Pharmacology. Encyclopedia 2021, 1, evolving techniques used to clarify its phylogeny [1].