Molecular Characterization of Unusual G10P[33], G6P[14] Genomic Constellations And
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Pune District Geographical Area
73°20'0"E 73°30'0"E 73°40'0"E 73°50'0"E 74°0'0"E 74°10'0"E 74°20'0"E 74°30'0"E 74°40'0"E 74°50'0"E 75°0'0"E 75°10'0"E PUNE DISTRICT GEOGRAPHICAL AREA To war a ds K ad (MAHARASHTRA) aly nw an- ha Dom m bi ra vali B P ds imp r a a l ¤£N g w H a o -2 T 19°20'0"N E o KEY MAP 2 2 n N Jo m 19°20'0"N g a A e D CA-01 TH THANE DINGORE 46 H CA-02 # S ta OTUR o Ma # B n JUNNAR s CA-03 ik AHMADNAGAR /" rd Doh D a ± CA-04 am w PUNE GEOGRAPHICAL o AREA (MNGL) TO BE CA-10 EXCLUDED FROM PUNE T DISTRICT GEOGRAPHICAL AREA UMBRAJ 0 # -5 CA-01 H N£ CA-05 DHALEWADI TARF HAVELI ¤ CA-09 CA-11 # Y ed ALE gaon Re T servoir Lake # ow 2 CA-06 22 a CA-08 H- r 19°10'0"N d RAJURI N s RAIGARH # £¤ T 19°10'0"N ak CA-07 CA-12 #NARAYANGAON #BORI BK. li D ho CA-13 ke Dim WARULWADI BELHE sh SOLAPUR bhe # w SATARA Da # S a m H r 5 1 KALAMB Total Population within the Geographical Area as per Census 2011 # T ow 46.29 Lacs (Approx.) GHODEGAON ar Total Geographical Area (Sq KMs) No. of Charge Areas ds S /" CA-02 H 1 Sh 14590 13 12 MANCHAR (CT) iru WADA r # .! Charge Area Identification Taluka Name C CA-01 Junnar 19°0'0"N ha CA-02 Ambegaon sk 19°0'0"N am an D CA-03 Khed a m CA-04 Mawal CA-05 Mulshi S PETH H 5 # CA-06 Velhe 4 i G d CA-07 Bhor h a T od Na o d w CA-08 Purandhar i( e w R CA-03 i n KADUS v CA-09 Haveli a e K a # r u r v ) k CA-10 Shirur d a d A s i G R CA-11 Daund N RAJGURUNAGAR i s H v e d a CA-12 Baramati /" r r v a M i w CA-13 Indapur M Wa o d i A v T u H 54 a le Dam S 62 18°50'0"N m SH D N SHIRUR 18°50'0"N b £H-5 ¤0 N a /" i CA-04 #DAVADI AG #KENDUR LEGEND KHADKALE -

MAHATMA PHULE MAHAVIDYALAYA Hon
OUR INSPIRATION ADVISORY COMMITTEE Hon. Sharadchandraji Pawar Hon. Dr. Nitin R. Karmalkar President, Vice Chancellor, Savitribai Phule University, Pune Dr. N. S. Umrani gm{dÌr~mB© ’w$co Rayat Shikshan Sanstha, Satara nwUo {dÚmnrR> Pro-Vice Chancellor, Savitribai Phule Uni., Pune Rayat Shikshan Sanstha's Hon. Prin. Dr. Arvind Burungale PATRONS S. M. Joshi, College, Hadapsar, Pune MAHATMA PHULE MAHAVIDYALAYA Hon. Dr. Anil Patil Hon. Prin. Dr. Manjushri Bobade PIMPRI-WAGHERE, PUNE - 411 017. Chairman, Rayat Shikshan Sanstha, Satara Dr. Babasaheb Ambedkar Mahavidyalaya, Aundh, Hon. N. D. Patil Pune Re-accredited by NAAC (Third Cycle) with ‘A’ Grade, CGPA : 3.16 Ex. Chairman and Member, Managing Council, Hon. Prin. D.D. Patil : 020-27412007 E-mail : [email protected] Rayat Shikshan Sanstha, Satara. Radhbai Kale Mahila Mahavidyalaya, A' Nagar Department of Psychology, Hon. Prin. Dnyandev Mhaske Hon. Prin. Dr. Bhausaheb Karale Physical Education and Economics Secretary, Rayat Shikshan Sanstha, Satara Jivajirao Shinde Mahavidayalaya, Shrigonda, A' Hon. Prin. Dr. Vijaysingh Sawant Nagar Organize Joint Secretary, Rayat Shikshan Sanstha, Satara Hon. Prin. Dr. Mohan Rajmane Two Day State Level Workshop on Shri. Vilas Mahadik S.G.M. College, Karad, Satara. Hon. Prin. Dr. Nanasaheb Gaikwad Joint Secretary, Rayat Shikshan Santha, Satara STRESS MANAGEMENT Hon. Prin. Dr. Arun Andhale Annasaheb Awate College Manchar, Pune Hon. Prin. Dr. K. G. Kanade, YCIS, Satara AT THE WORK PLACE Auditor, Rayat Shikshan Santha, Satara Dr. Deepak Mane Hon. Ajitdada Pawar th th Director, Board of Sports & Physical Education, 16 to 17 January 2019 Member, Managing Council, SPPU, Pune Rayat Shikshan Sanstha, Satara. Dr. Narendra Deshmukh Hon. -

Under Government Orders
(Under Government Orders) BOMBAY PlUNTED AT THE GOVERNMENT CENTlUI. PRESS )btainable from the Government Publications Sales Depot, Institute of Science ' Building, Fort, Bombay (for purchasers in Bombay City); from the Government Book Depot, Chami Road Gardens, Bombay 4 (for orders from the mofussil) or I through the High Commissioner for India, India House, Aldwych, London. W.C.2 . or through any recognized Bookseller. Price-Re. 11 Anna.s 6 or 198. 1954 CONTENTS 1lJ. PAGB PREFACE v GENERAL INTRODUCTION • VII-X MAP. PART I. CHAPTER 1 : PHYSICAL FEATURES .urn NATURAL REsOURCES- 1 Boundaries and Sub-Divisions 1 ; ASpects 2 ; Hills 4 ; River Systems 6; Geology 10 ; Climate 11; Forests 20; Fauna 24 ; Birds 28; Fish 32; Snakes 37. PART n. CHAPTER 2: ADMINISTRATIVE HISTORY- ,(1 Hindu Period ~90 B.C.-1295 A.D.) 41; Muslim Period (1295-1720) 43; Maratha Period \1720-1818) 52; British Period (1819-1947) 59. PART m. CIIAPTE~ 3: TIm, ~OPLE .AND Tm:m CULTURE-.- 69 Population' (1951 Census) 69; Food 75; Houses and Housing 76; Dress 78; Ornaments 21 ; Hindu CUstoms 82 ; Hindu Religious Practices 120;. Gaines 137; Dances 141; Akhadas or TaIims 145; ·Tamasha 146; Bene Israels'147; Christians 150; Muslims 153 ~ People from Tamil Nad 'and Kerala 157; Sindhi Hindus, 159. P~T IV....iECONOMIC ORGAN1ZAT~ON. CHAPTER 4: GENERAL ECONOMIC SURVEY .. 163 CHAPTER 5 : A~CULTUllE- 169 Agricultural .Popillation 169.; Rainfall 172; 'Agricultural Season 173; Soils 174; Land Utilization 177 j Holdings 183; Cereals 191; Pulses 196; Oil-Seeds 199; Drugs and Narcotics 201; Sugarcane 202; Condiments and Spices 204; Fibres 206; Fruits and Vegetables 207; AgricUltural. -

Ffir*Rfi, Got Irext Strr+ Qerpift=[ Yrflr5.{Ur, Gut6ftilr Uift, F4{D?Frfr HOSPITAL MISE ALTOT MENT of IEMDES IVIR on 6-04-2021PUN - Dtstrtc'l
x'.fq.6, .r+. en lhtfue-q qffilr ! e/Q o q q ffiElql,Frq,got ffrt*,. ?Q/x/qoqq qfitqffi fuEq :- Hftrtr{ ifuE-qT qreqrc{r-{f,. liqrt '- l) qr. sTr{ffi , Bl=r H si}qq urTlvq *'+ qiq*oqr.m.qlfqerJHfisqtr q, q. qfrq{H/q. 1- R k{is- q/x/R o R t) qr qrqffie qr mreffim e+rtvr *. fe.HT. ien.q /ffi /qst /tott.kqi6 q\/x/RoRt. i) qT *lqidqmslfr qr qrqffie entvr m. fq.mr. /en.q /ffi /q\er /totq.kim U/x/QoRq. x) ur.o1g6, sr=r.r eiiqq wnaq qtffif, qr F. .No. Remdesivir nr 4/ $' 202t/08 frqim qi/x/Ro?l e+re k{i6. Rq/x/?oRt n-S gri ffi srw Art'er Hnr+{ tffi Fru1?1-q F-orq eraq Hi-q-d-qT qffi s-{u-{Trd sTrA-f,1 GG. ilft {GiRrd +ii{s eit qurqqhr q E{tfoqm 3Tr*@T lffi ertwr gwd-Et-{ 1q-n ei++q{ HGFra. tq-gfiselr idErH rTrsfli FR+d ffi--@T qr+ qrq q;sq qr*o qt{IEkT qi"kflEt Req qwrr ilE}.ffi e-snql tffi , ztarzrd, wq{ Elw{ rT6t qrfr qerf,r Frulrei+ wrnr-{ q qr$q' m qirt fuH sTrt {Eiiqn *t+s FrutT-f,qirt vflqT et}qq {rar qTw q'-FT ndffirqt Fione-qrqr d'er t-g r{ v& mmn* qrfrrsrrd e vrPr$f, q-ffiti qrs.F ffi {K{ q-rui qFi[{s' qTt qurc-d-qt qktsmr swi qrger Bgirt eftqqi \Fl.q fuf,{q v<t qTM tfiisrs Eq-frn q eil-s*tq / atsfi wro +-wti ent *tfue-t q a{ko{refiqd o.rq uruntfrdr$T Tdr{' yqr: aml'q fur, rcmrrwnfuor, qrrqrfufi, rr6Eit, +*q, urqkflq, t.nf+ors, srq E[ oitqtl q$nff{, qfr€{ FFTrfi FdPdq enenr<iqr orRr*,rff e offi mffi ro z*^qtewr*d *qoqd fqtw qt. -

Maharashtra State Electricity Distribution Company Ltd. Feeder Interruption Details of Planned Outages for MAY-16 Report Date : Jun 16, 2016
Maharashtra State Electricity Distribution Company Ltd. Feeder Interruption Details of Planned Outages for MAY-16 Report Date : Jun 16, 2016 Zone Circle Division Subdivision Substation Feeder Town Interruption Start Date Interruption End Interruption Period (Days - Total DTCs on Reason of Time Date Time Hr-Min-Sec) Feeder Interruption AKOLA AKOLA AKOLA URBAN 4275- AKOLA U-I S/DN 024029- 33 KV 204- 11 KV Cotton 101- AKOLA 15-MAY-2016 06:00:00 15-MAY-2016 08:51:00 0 - 02-51-00 24 DLS (Discrete Load ZONE CIRCLE DIVISION Mohata Sub Station Market Shedding) AKOLA AKOLA AKOLA URBAN 4275- AKOLA U-I S/DN 024029- 33 KV 204- 11 KV Cotton 101- AKOLA 15-MAY-2016 09:30:00 15-MAY-2016 10:45:00 0 - 01-15-00 24 DLS (Discrete Load ZONE CIRCLE DIVISION Mohata Sub Station Market Shedding) AKOLA AKOLA AKOLA URBAN 4275- AKOLA U-I S/DN 024048- 33/11 KV 205- 11 KV 101- AKOLA 15-MAY-2016 06:00:00 15-MAY-2016 09:00:00 0 - 03-00-00 17 Planned loading ZONE CIRCLE DIVISION Shivaji Nagar Sub- Ganesh Nagar shedding station Feeder AKOLA AKOLA AKOLA URBAN 4275- AKOLA U-I S/DN 024048- 33/11 KV 205- 11 KV 101- AKOLA 15-MAY-2016 15:30:00 15-MAY-2016 17:09:00 0 - 01-39-00 17 Planned loading ZONE CIRCLE DIVISION Shivaji Nagar Sub- Ganesh Nagar shedding station Feeder AKOLA AKOLA AKOLA URBAN 4592- AKOLA U-III 024030- VIDYUT 202- 101- AKOLA 15-MAY-2016 00:00:00 15-MAY-2016 00:00:00 0 - 00-00-00 50 Planned loading ZONE CIRCLE DIVISION S/DN BHAVAN S/S RAMDASPETH shedding AKOLA AKOLA AKOLA URBAN 4592- AKOLA U-III 024030- VIDYUT 202- 101- AKOLA 15-MAY-2016 06:00:00 15-MAY-2016 -

A Study of Basic Amenities in Tribal Area of Ambegaon Taluka with Special Reference to Electricity and Telecommunication
Pramana Research Journal ISSN NO: 2249-2976 A Study of Basic Amenities in Tribal Area of Ambegaon Taluka with Special Reference to Electricity and Telecommunication Prof. Mayur Chikhale*1, Assistant Professor, Adhalraon Patil Institute of Management and Research, Landewadi, Manchar, Pune | Email- [email protected] Dr. Dhananjay Pingale, PhD Associate Professor, Adhalraon Patil Institute of Management and Research, Landewadi, Manchar, Pune | Email- [email protected] Abstract Ambegaon Tehsil is classified as tribal area in Pune District and having 23 per cent of land covered the forest, providing home to Hindu Mahadev Koli peoples (tribal) in the vicinity. The present study put its efforts on understanding the level of basic amenities provided to these tribal peoples in terms of transportation, drinkable water and telecommunication facility. The paper studied 15 villages of Ambegaon Tehsil and investigated opinions of 51 tribal peoples. Ultimately, it has observed that, transport facility is not sufficiently provided and arrangement of alternate energy source for cooking needs to be provided. These are the post research policy suggestions given based on the investigation made in this research study. Keywords: Basic Amenities, Rural Area of Ambegaon Tehsil, Electricity and Telecommunication Introduction It is highly observed that the urban area peoples and rural area peoples have very differential lifestyles as well as their problems. This has reflected in considering the present research for understanding on the pain areas for suffering a lot in regards with the basic needs of the rural life. The present study focuses on basic amenities or may be called as basic needs for 21st century citizens, namely, (a) transportation facility available in the vicinity, (b) market accessibility from the residential place, (c) drinkable water facility, and (d) telecommunication facility available. -

State District Branch Address Centre Ifsc Contact1 Contact2 Contact3 Micr Code
STATE DISTRICT BRANCH ADDRESS CENTRE IFSC CONTACT1 CONTACT2 CONTACT3 MICR_CODE Alandi Markal Road, Mr. MAHARAS Taluka-Khed, District- Gajanan 02135- HTRA PUNE Alandi Pune, 412105 ALANDI RSBL0000014 Choudhari - 233285 411164320 D.R. Kakade Plaza, Mr. MAHARAS Alephata, Taluka- NARAYANG Ramesh 02132- HTRA PUNE Alephata Junnar, 410501 AON RSBL0000013 Ghumatkar - 263085 319,320 Pune-Nashik Highway, Rajgurunagar Tal. MAHARAS Khed Dist Pune RAJGURUN Mr. Ashok 02135- HTRA PUNE Bajar Peth 410505 AGAR RSBL0000003 Oswal 222107 411164301 Agarwal Building, Gawali Nagar, Bhosari, MAHARAS Taluka-Haweli, District- Mr. Dilip 020- HTRA PUNE Bhosari Pune, 411039 PUNE RSBL0000007 Malaghe - 27110210 411164281 634, Shri Vidya- Bhavan, Budhwar MAHARAS Peth, District-Pune, Mr. Prasad 020- HTRA PUNE Budhwar Peth 411002 PUNE RSBL0000009 Belhekar - 24463132 411164294 Aiswarya Icon, Ambethan Chowk, MAHARAS Chakan, Taluka-Khed, Mr. Ankush 02135- HTRA PUNE Chakan District-Pune, 410501 CHAKAN RSBL0000004 Kohinkar - 249056 411164283 Agrasen Chowk, Pune- Mumbai Road, Dehu Mr. MAHARAS Road, Taluka-Haweli, Rajendra 020- HTRA PUNE Dehu Road District-Pune, 412101 DEHU ROAD RSBL0000008 Katariya - 27673011 411164284 319/320 Pune-Nashik Highway Rajgurunagar, Tal. Mrs. MAHARAS Khed, Dist Pune RAJGURUN Kakade Mr. Amrut 02135- HTRA PUNE Head Office 410505 AGAR RSBL0000001 Jyotsna Takalakar 222085 Grampanchayat Building, Kadus, MAHARAS Taluka-Khed, District- RAJGURUN Mr. Vijay 02135- HTRA PUNE Kadus Pune, 412404 AGAR RSBL0000005 Satkar - 282244 Gram Panchayat Complex, Shivali Chowk, Manchar, MAHARAS Taluka-Ambegaon, Mr. Balu 02133- HTRA PUNE Manchar District-Pune, 410503 MANCHAR RSBL0000010 Gholap - 223716 Chakan Talegaon Road, A/P Mahalunge- MAHARAS Ingale, Taluka-Khed, Mr. Sunil 02125- HTRA PUNE Mhalunge-Ingale District-Pune, 410501 CHAKAN RSBL0000012 Pansare - 259059 411164319 Gramvaibhav Complex, Junnar Road, Narayangaon, MAHARAS Taluka-Ambegaon, NARAYANG Mr. -

Vffiaqffi?Qiardffiri+Wtq Frfuil-Q-+. Ewft Qwffirr Onfu Qdtrn Dffu Qiqr Qr Atqfdqtqr*A Ffwn )Qdfud\T I+Qw Dt7wqr Gadr Aaw6 Qffiatar Mq Qmdt Gdgqnr T Dar Ilv Dfu
m.fu.m./* en /*tfus- q qisTfr/e o/q o q q iwerf lr*rn u,T qicrq, gui fui-*^ Xo/oq/qoq1 qfrErm fqqq :- tqtfi$qh- i+wrqT qreqqrcffi. xiel :- q) qr. eirTm, Gl=r q e\qE q{rml-{ fi q-fiqrs-er.m.o}f+st-7ffiq Ef,qqm71o1-tq, kqis q/xho lt. qI m-rqlmqrs-*m qr mrqffio BrT*gl^ m. fq.qr. /en.q /mrfu /qsq q'frqi*. q\ixlRo?q. *ffieqq'rqffio 3lTkrm. fq.*.r. /en.q ffi /qrgr ffi .fr{i6 tq/x/?oRq. 6i,.irfl*-a&lf,".* Br=I qiffiiT rrr +r.etlWi tt.eTr-{ffi, E stqE v{TRFI F. .No. Remdesivir tLZ 47 2t/ 0B tfliq. qi/x/Ro?q €IrE kqt*. Qo/oq/QoQ{ ffi g't lW qril fld-eT q11q Hfir*{ E++qrq e +{ qTzqr€rat BqtreT ewd-€r q11 ffi{ E+flH erqn qsq lotQl HfiH-+{ Effi srunfiq qrqlmTui fr65q qre FtEd-qT *-grqrf, Gilffi eTrt H{t {ciftrf, s}ks s,.qeqiqt Hi+ =ndTqEk qqtkuqTd 3rT-+eT ffi E stqq Twdrqr I+m .ut+*Eq f,rFfl-a iqtFr*{ ii+wH yrFF{fi q-sq qrfl-qf, fn+f, ++eT q{ri ma q-}6 . qffi} srq q?iqm qlE1ffi qsR+ fti'nt , zrarem, eq{ EtqT{ Tdr qT-+ Rtdr Frurcrq qflRl-{ E qrss-fu+fr qi-fi irt+ qTt {cifrId ++s Frundqifr sfl"fi 3fiqq q6 qTH sF=T nnffi glqrtrqr€qr +c{ ts q{ s61 ffi mftrq;Rq{ q HftrFd qffiti oU ertreq* qrs.h- ffi qrcr *-{0} s+rw+o ent qrqwirqt EkRcIfi sF=T qrsEr-ffiE1 eiqt{r+ g+q kil{q vqr sTrffi frrtffiq g+fro e vtrqqlq / qlsfr ErTd strt-i eTrt trqd Er*f, ewfr flq Frqvlfl€ 3Trmt eTrt 6\,61fua6qda Fluti+ iM*. -
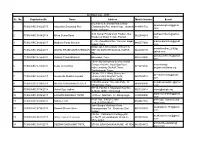
Architect List - 2019 Sr
Architect List - 2019 Sr. No. RegistrationNo Name Address Mobile Number E-mail 642,Flat no 9, Snehal Park,Behind splusadesigners@gmail. 1 PCMC/ARC/0652/2017 Adityasinh Dayanand Patil Chandrakant Patil Heart Hosp. Jawahar 8149991732 com Nagar, Kolhapur. A/16 Kumar Priydarshan Pashan, Sus subhaarchitects@yahoo. 2 PCMC/ARC/0438/2018 Milind Subha Saraf 9822554283 Road,near Balaji Temple Pashan com C - 16, Jivandhara Soc. Yamuna nagar, madhuraarchitect@gmail. 3 PCMC/ARC/0692/2017 Madhura Parag Merukar 9860577999 Nigadi- Pune com SHOP NO 1,SHIVANJALI HEIGHTS anandkhedkar_2000@ 4 PCMC/ARC/0562/2017 ANAND PRABHAKAR KHEDKAR BEHIND BORATE SANKUL KARVE 9822400439 yahoo.com RD. sucratuarchitects@gmail. 5 PCMC/ARC/0725/2018 Siddesh Pravin Bhansali Bibvewadi, Pune. 9028783400 com 1901/1902 Drewberry Everest World Complex Kolshet Road,Opp Bayer kedar.bhat@ 6 PCMC/ARC/0768/2018 Kedar Arvind Bhat 9819519195 India Company Dhokali,Thane, srujanconsultants.org Sandozbaugh Thane. Flat no. 102 J- Wing, Survey no directionnextds@gmail. 7 PCMC/ARC/0682/2017 Amannulla Shabbir Inamdar 5A/2A,212B/2, Mayfair Pacific, 9657009789 com Kondhawa Khurd Pune,NIBM C/O-AR.Laxman Thite Sita Park, 18, milind.laxmanthite@gmail 8 PCMC/ARC/0399/2018 MILIND RAMCHANDRA PATIL 8408880898 Shivajinagar, Pune .com RH 55, Flat No 8, Nityanand Hsg Soc, 9 PCMC/ARC/0718/2018 Vishal Vijay Jadhav 9923128414 [email protected] G-Block, MIDC, Chinchwad datta.laxmanthite@gmail. 10 PCMC/ARC/0532/2017 LAXMAN SADASHIV THITE 1st Floor, Sita Park, 18, Shivajinagar, 8408880890 com PLOT NO - 390,SECTOR archetype_associates@ 11 PCMC/ARC/0074/2017 Nafisa A Kazi 9922007885 27/A,PCNTDA,NIGDI gmail.com Janiv Bangla Malshiras Road swapnilgirme173@gmail. -

List of Police Station Wise Judicial Officers in Pune District
List of police station wise Judicial Officers in Pune District . Sr. Office Phone Name of the Police station Name of the Court No. No. attached to the court 2 3 4 Shri. M.S. Agrawal, 1 26340885 Shivajinagar Addl. C.J.M., Court, Pune. Smt. J. S. Kelkar, Samartha, Kothrud and 2 25534796 JMFC Court No. 1, Pune. Uttamnagar Smt. R. K. Bafna/Bhalgat, 3 25534796 Faraskhana & Dattawadi JMFC Court No. 2, Pune. Smt. T.M. Nirale, Bandgarden,Haveli,Koregaon 4 25534796 JMFC Court No. 3, Pune park & Warje Malwadi Sahakar Nagar, Bibvewadi, Shri. P. S. Jondhale, 5 25534796 Visharambug & Bharti JMFC Court No. 4, Pune Vidyapeeth Shri. V. I. Shaikh, Yerwada, Chandannagar & 6 25534796 JMFC Court No.5, Pune Velha Smt. A.S.Mujumdar, Hinjawadi, Lonikand and 7 25534796 JMFC Court No.6, Pune Cyber Crime Pune City. Shri. S. B. Ganapa, 8 Loni Kalbhor and Paud JMFC Court No. 7, Pune Smt. S. R. Patil, 9 Swargate and Market Yard JMFC Court No. 8, Pune Smt.A.V.Mohite, 10 25534796 chaturshringi pol stn JMFC Court No.9, Pune Smt. S.K.Khan, 11 25534796 Vimantal 12th Jt.JMFC, Pune. Shri. M. A. Shaikh, 12 25534796 Khadak & Alankar 13th Jt.JMFC, Pune. Shri. S. B. Patil, 13 25534796 Deccan, Sinhagadroad, A.T.S. (A.C.) Court, Pune. Shri. N.K.Kharade, 14 25534796 All Traffic branches at Pune (M.V.) Court, Pune. Shri. K. C. Rajput, 15 26137247 Hadapsar, Lashkar & Mundhwa JMFC.(Cant) Pune. Shri. A. S. Deshpande, 16 25534796 Kondhwa Jt.JMFC (Cant) Pune. Smt. R.S.Patil, 17 25534796 Wanwadi 2nd Jt.JMFC (Cant) Pune. -

Reimagining Water Infrastructure in Its Cultural Specificity
Reimagining Water Infrastructure in its Cultural Specificity Case of Pune, INDIA. Manas Rajendra Marathe Supervisors Prof. Dr-Ing. Annette Rudolph-Cleff Prof. Dr Gerrit Jasper Schenk Fachgebiet Entwerfen und Stadtwicklung Fachbereich Architektur 2019 REIMAGINING WATER INFRASTRUCTURE IN ITS CULTURAL SPECIFICITY Case of Pune, INDIA. Genehmigte Dissertation zur Erlangung des akademischen Grades Doktor der Ingenieurwissenschaften (Dr-Ing.) von M.Sc. Manas Rajendra Marathe aus Pune, Indien. Graduiertenkolleg KRITIS 1. Gutachter: Prof. Dr-Ing. Annette Rudolph-Cleff 2. Gutachter: Prof. Dr Gerrit Jasper Schenk Tag der Einreichung: 11-09-2019 Tag der Prüfung: 21.10.2019 Fachgebiet Entwerfen und Stadtwicklung Fachbereich Architektur (FB 15) Technische Universität Darmstadt L3 01 El- Lissitzky Straße 1 64287 Darmstadt URN: urn:nbn:de:tuda-tuprints-92810 URI: https://tuprints.ulb.tu-darmstadt.de/id/eprint/9281 Published under CC BY 4.0 International https://creativecommons.org/licenses/ Darmstadt, October 2019. Cover page: Photo of Barav at Loni Bhapkar, Pune. Erklärung zur Dissertation Hiermit versichere ich, die vorliegende Dissertation ohne Hilfe Dritter nur mit den angegebenen Quellen und Hilfsmitteln angefertigt zu haben. Alle Stellen, die aus Quellen entnommen wurden, sind als solche kenntlich gemacht. Diese Arbeit hat in gleicher oder ähnlicher Form noch keiner Prüfungsbehörde vorgelegen. Darmstadt, den 11-09-2019. ________________________ (Manas Rajendra Marathe) Acknowledgements The culmination of this dissertation would not have been possible without the help and support of many people and institutions. Firstly, I express my sincere gratitude towards my Supervisor Prof. Dr-Ing. Annette Rudolph Cleff and my Co-Supervisor Prof. Dr Gerrit Jasper Schenk for their constant encouragement, guidance and wholehearted support. -

Pune Cdap.Pdf
PREFACE The process of planned economic development in India began with the launching of the First Five Year Plan in 1951 and currently India is in the 12th Five Year Plan (2012-13 to 2016- 2017). The main objective of policy makers is to promote growth with social justice. During the Eleventh Plan period, the agricultural sector experienced a miniscule growth rate of 3.64 per cent per annum. Indian agriculture is presently at cross roads and one of the major challenges is to reverse deceleration in agricultural growth rates so as to successfully achieve a higher broad based growth. In view of the above, a special Additional Central Assistance Scheme -Rashtriya Krishi Vikas Yojna (RKVY) which is a State Plan scheme administered by the Union Ministry of Agriculture was conceived. The main purpose of the scheme is to supplement state specific strategies with a view to rejuvenate agriculture. The pattern of funding under this scheme is 100 percent Central grant. In order to avail of funds under RKVY, each district is entrusted with the task of preparing a comprehensive district agricultural plan. Accordingly, this plan was prepared for Pune district. The Plan revealed that the city has suitable infrastructure and conducive climate for high value agriculture. Floriculture is also coming up in a big way. Similarly dairy development and poultry have huge potential in the district. As the city is close to Mumbai and well connected by road, rail and air to other major cities there is a ready market available for consumption of agricultural goods, processed goods, dairy and poultry products.