Cell Cycle/DNA Damage
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Sequence-Selective Recognition of Double-Stranded RNA And
Downloaded from rnajournal.cshlp.org on October 8, 2021 - Published by Cold Spring Harbor Laboratory Press Hnedzko et al. Sequence-Selective Recognition of Double-Stranded RNA and Enhanced Cellular Uptake of Cationic Nucleobase and Backbone- Modified Peptide Nucleic Acids Dziyana Hnedzko1,*, Dennis W. McGee,2 Yannis A. Karamitas1 and Eriks Rozners1,* Departments of 1 Chemistry and 2 Biological Sciences, Binghamton University, The State University of New York, Binghamton, New York 13902, United States. * To whom correspondence should be addressed. Tel: +1-607-777-2441; Fax: +1-607-777- 4478; Email: [email protected] and [email protected] Running Tittle: RNA recognition and cellular uptake of PNA 1 Downloaded from rnajournal.cshlp.org on October 8, 2021 - Published by Cold Spring Harbor Laboratory Press Hnedzko et al. ABSTRACT Sequence-selective recognition of complex RNAs in live cells could find broad applications in biology, biomedical research and biotechnology. However, specific recognition of structured RNA is challenging and generally applicable and effective methods are lacking. Recently, we found that peptide nucleic acids (PNAs) were unusually well suited ligands for recognition of double-stranded RNAs. Herein, we report that 2-aminopyridine (M) modified PNAs and their conjugates with lysine and arginine tripeptides form strong (Ka = 9.4 to 17 × 107 M-1) and sequence-selective triple helices with RNA hairpins at physiological pH and salt concentration. The affinity of PNA-peptide conjugates for the matched RNA hairpins was unusually high compared to the much lower affinity for DNA hairpins of the same 7 -1 sequence (Ka = 0.05 to 0.11 × 10 M ). The binding of double-stranded RNA by M-modified 4 -1 -1 PNA-peptide conjugates was a relatively fast process (kon = 2.9 × 10 M s ) compared to 3 -1 -1 the notoriously slow triple helix formation by oligodeoxynucleotides (kon ~ 10 M s ). -

Human Cytogenetics Prenatal Diagnostics
Cytogenetics Human Cytogenetics Prenatal Diagnostics Optimized Medium for Culture and Genetic Analysis of Human Amniotic Fluid Cells BIOAMF-1 and Chorionic Villi ( CV ) Samples Basal Medium and Supplement Chromosome Karyotyping was first developed in the BIOAMF-1 is designed for the primary culture of field of Cytogenetics. human amniotic fluid cells and chorionic villi (CV) The basic principle of the method is the preparation samples in both open (5% CO2) and closed systems. of chromosomes for microscopic observation by The medium allows rapid growth of amniocytes or arresting cell mitosis at metaphase with colchicine and treating the cells with a hypotonic solution. This chorionic villi for use in karyotyping. is followed by regular or fluorescent staining of the No supplementation with serum or serum- chromosomes, which are then tested with the aid of a substitutes is necessary. microscope and computer programs to arrange and The medium consists of two components: basal identify the chromosomes for the presence of genetic medium and frozen supplements. abnormalities. In principle, this method enables the identification Instructions for Use of any abnormality - excess chromosomes or For the preparation of 500ml complete medium, use chromosome deficiency, broken chromosomes, 01-190-1A with 01-192-1E. or excess genetic material (as a result of a For the preparation of 100ml complete medium, use recombination process). 01-190-1B with 01-192-1D. Clinical cytogenetics laboratories use this method Thaw the BIOAMF-1 Supplement by swirling in a with amniotic fluid, chorionic villi, blood cells, skin cells, and so on, which can be cell cultured to obtain 37ºC water bath, and transfer the contents to the mitotic cells. -
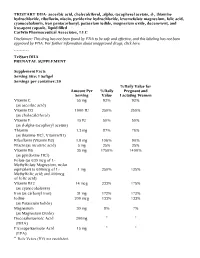
TRISTART DHA- Ascorbic Acid, Cholecalciferol, .Alpha
TRISTART DHA- ascorbic acid, cholecalciferol, .alpha.-tocopherol acetate, d-, thiamine hydrochloride, riboflavin, niacin, pyridoxine hydrochloride, levomefolate magnesium, folic acid, cyanocobalamin, iron pentacarbonyl, potassium iodide, magnesium oxide, doconexent, and icosapent capsule, liquid filled CarWin Pharmaceutical Associates, LLC Disclaimer: This drug has not been found by FDA to be safe and effective, and this labeling has not been approved by FDA. For further information about unapproved drugs, click here. ---------- TriStart DHA PRENATAL SUPPLEMENT Supplement Facts Serving Size: 1 Softgel Servings per container: 30 %Daily Value for Amount Per %Daily Pregnant and Serving Value Lactating Women Vitamin C 55 mg 92% 92% (as ascorbic acid) Vitamin D3 1000 IU 250% 250% (as cholecalciferol) Vitamin E 15 IU 50% 50% (as d-alpha-tocopheryl acetate) Thiamin 1.3 mg 87% 76% (as thiamine HCl, Vitamin B1) Riboflavin (Vitamin B2) 1.8 mg 106% 90% Niacin (as nicotinic acid) 5 mg 25% 25% Vitamin B6 35 mg 1750% 1400% (as pyridoxine HCl) Folate (as 630 mcg of L- Methylfolate Magnesium, molar equivalent to 600mcg of L- 1 mg 250% 125% Methylfolic acid; and 400mcg of folic acid) Vitamin B12 14 mcg 233% 175% (as cyanocobalamin) Iron (as carbonyl iron) 31 mg 172% 172% Iodine 200 mcg 133% 133% (as Potassium Iodide) Magnesium 30 mg 8% 7% (as Magnesium Oxide) Docosahexaenoic Acid 200mg * * (DHA) Eicosapentaenoate Acid 15 mg * * (EPA) * Daily Values (DV) not established. OTHER INGREDIENTS: Gelatin (bovine), Glycerin, Purified Water, Yellow Bees Wax, Caramel Powder, Soy Lecithin, Natural Orange Flavor, Ethyl Vanillin. Contains: Soy and Fish TriStart DHA™ Softgel capsules are dye free, lactose, gluten and sugar free. -

SERS and MD Simulation Studies of a Kinase Inhibitor Demonstrate the Emergence of a Potential Drug Discovery Tool
SERS and MD simulation studies of a kinase inhibitor demonstrate the emergence of a potential drug discovery tool Dhanasekaran Karthigeyana,1, Soumik Siddhantab,1, Annavarapu Hari Kishorec,1, Sathya S. R. R. Perumald, Hans Ågrend, Surabhi Sudevana, Akshay V. Bhata, Karanam Balasubramanyama, Rangappa Kanchugarakoppal Subbegowdac,2, Tapas K. Kundua,2, and Chandrabhas Narayanab,2 aTranscription and Disease Laboratory, Molecular Biology and Genetics Unit, bLight Scattering Laboratory, Chemistry and Physics of Materials Unit, Jawaharlal Nehru Centre for Advanced Scientific Research, Jakkur, Bangalore 560064, India; cDepartment of Studies in Chemistry, University of Mysore, Manasagangotri, Mysore 570006, India; and dDepartment of Theoretical Chemistry and Biology, School of Biotechnology, KTH Royal Institute of Technology, Roslagstullsbacken 15, SE-114 21 Stockholm, Sweden Edited by Michael L. Klein, Temple University, Philadelphia, PA, and approved May 30, 2014 (received for review February 18, 2014) We demonstrate the use of surface-enhanced Raman spectroscopy be screened for therapeutic applications. This paper provides a (SERS) as an excellent tool for identifying the binding site of small prelude to this development. This finding also facilitates the molecules on a therapeutically important protein. As an example, developing field of tip-enhanced Raman spectroscopy for im- we show the specific binding of the common antihypertension aging the small molecule interactions for in vitro and in vivo drug felodipine to the oncogenic Aurora A kinase protein via applications. A completely developed SERS–MD simulation hydrogen bonding interactions with Tyr-212 residue to specifically combination with adequate help from the structure of the pro- inhibit its activity. Based on SERS studies, molecular docking, tein may help converge potential small molecules for therapeutic molecular dynamics simulation, biochemical assays, and point applications and reduce the time for drug discovery. -

Clinical Candidates Targeting the ATR–CHK1–WEE1 Axis in Cancer
cancers Review Clinical Candidates Targeting the ATR–CHK1–WEE1 Axis in Cancer Lukas Gorecki 1 , Martin Andrs 1,2 and Jan Korabecny 1,* 1 Biomedical Research Center, University Hospital Hradec Kralove, Sokolska 581, 500 05 Hradec Kralove, Czech Republic; [email protected] (L.G.); [email protected] (M.A.) 2 Laboratory of Cancer Cell Biology, Institute of Molecular Genetics of the Czech Academy of Sciences, Videnska 1083, 142 00 Prague, Czech Republic * Correspondence: [email protected]; Tel.: +420-495-833-447 Simple Summary: Selective killing of cancer cells is privileged mainstream in cancer treatment and targeted therapy represents the new tool with a potential to pursue this aim. It can also aid to overcome resistance of conventional chemo- or radio-therapy. Common mutations of cancer cells (defective G1 control) favor inhibiting intra-S and G2/M-checkpoints, which are regulated by ATR–CHK1–WEE1 pathway. The ATR–CHK1–WEE1 axis has produced several clinical candidates currently undergoing clinical trials in phase II. Clinical results from randomized trials by ATR and WEE1 inhibitors warrant ongoing clinical trials in phase III. Abstract: Selective killing of cancer cells while sparing healthy ones is the principle of the perfect cancer treatment and the primary aim of many oncologists, molecular biologists, and medicinal chemists. To achieve this goal, it is crucial to understand the molecular mechanisms that distinguish cancer cells from healthy ones. Accordingly, several clinical candidates that use particular mutations in cell-cycle progressions have been developed to kill cancer cells. As the majority of cancer cells have defects in G1 control, targeting the subsequent intra-S or G2/M checkpoints has also been extensively Citation: Gorecki, L.; Andrs, M.; pursued. -

Nuclear and Mitochondrial DNA Sequences from Two Denisovan Individuals
Nuclear and mitochondrial DNA sequences from two Denisovan individuals Susanna Sawyera,1, Gabriel Renauda,1, Bence Violab,c,d, Jean-Jacques Hublinc, Marie-Theres Gansaugea, Michael V. Shunkovd,e, Anatoly P. Dereviankod,f, Kay Prüfera, Janet Kelsoa, and Svante Pääboa,2 aDepartment of Evolutionary Genetics, Max Planck Institute for Evolutionary Anthropology, D-04103 Leipzig, Germany; bDepartment of Anthropology, University of Toronto, Toronto, ON M5S 2S2, Canada; cDepartment of Human Evolution, Max Planck Institute for Evolutionary Anthropology, D-04103 Leipzig, Germany; dInstitute of Archaeology and Ethnography, Russian Academy of Sciences, Novosibirsk, RU-630090, Russia; eNovosibirsk National Research State University, Novosibirsk, RU-630090, Russia; and fAltai State University, Barnaul, RU-656049, Russia Contributed by Svante Pääbo, October 13, 2015 (sent for review April 16, 2015; reviewed by Hendrik N. Poinar, Fred H. Smith, and Chris B. Stringer) Denisovans, a sister group of Neandertals, have been described on DNA to the ancestors of present-day populations across Asia the basis of a nuclear genome sequence from a finger phalanx and Oceania suggests that in addition to the Altai Mountains, (Denisova 3) found in Denisova Cave in the Altai Mountains. The they may have lived in other parts of Asia. In addition to the only other Denisovan specimen described to date is a molar (Deni- finger phalanx, a molar (Denisova 4) was found in the cave in sova 4) found at the same site. This tooth carries a mtDNA se- 2000. Although less than 0.2% of the DNA in the tooth derives quence similar to that of Denisova 3. Here we present nuclear from a hominin source, the mtDNA was sequenced and differed DNA sequences from Denisova 4 and a morphological description, from the finger phalanx mtDNA at only two positions, suggesting as well as mitochondrial and nuclear DNA sequence data, from it too may be from a Denisovan (2, 3). -

Hammerhead Ribozymes Against Virus and Viroid Rnas
Hammerhead Ribozymes Against Virus and Viroid RNAs Alberto Carbonell, Ricardo Flores, and Selma Gago Contents 1 A Historical Overview: Hammerhead Ribozymes in Their Natural Context ................................................................... 412 2 Manipulating Cis-Acting Hammerheads to Act in Trans ................................. 414 3 A Critical Issue: Colocalization of Ribozyme and Substrate . .. .. ... .. .. .. .. .. ... .. .. .. .. 416 4 An Unanticipated Participant: Interactions Between Peripheral Loops of Natural Hammerheads Greatly Increase Their Self-Cleavage Activity ........................... 417 5 A New Generation of Trans-Acting Hammerheads Operating In Vitro and In Vivo at Physiological Concentrations of Magnesium . ...... 419 6 Trans-Cleavage In Vitro of Short RNA Substrates by Discontinuous and Extended Hammerheads ........................................... 420 7 Trans-Cleavage In Vitro of a Highly Structured RNA by Discontinuous and Extended Hammerheads ........................................... 421 8 Trans-Cleavage In Vivo of a Viroid RNA by an Extended PLMVd-Derived Hammerhead ........................................... 422 9 Concluding Remarks and Outlooks ........................................................ 424 References ....................................................................................... 425 Abstract The hammerhead ribozyme, a small catalytic motif that promotes self- cleavage of the RNAs in which it is found naturally embedded, can be manipulated to recognize and cleave specifically -

Enbrace® HR DESCRIPTION: INGREDIENTS
EnBrace® HR with DeltaFolate ™ [1 NF Units] [15 mg DFE Folate] ANTI-ANEMIA PREPARATION as extrinsic/intrinsic factor concentrate plus folate. Prescription Prenatal/Vitamin Drug For Therapeutic Use Multi-phasic Capsules (30ct bottle) NDC 64661-650-30 Rx Only [DRUG] GLUTEN-FREE DESCRIPTION: EnBrace® HR is an orally administered prescription prenatal/vitamin drug for therapeutic use formulated for female macrocytic anemia patients that are in need of treatment, and are under specific direction and monitoring of vitamin B12 and vitamin B9 status by a physician. EnBrace® HR is intended for women of childbearing age who are – or desire to become, pregnant regardless of lactation status. EnBrace® HR may be prescribed for women at risk of depression as a result of folate or cobalamin deficiency - including folate-induced postpartum depression, or are at risk of folate-induced birth defects such as may be found with spina bifida and other neural tube defects (NTDs). INGREDIENTS: Cobalamin intrinsic factor complex 1 NF Units* * National Formulary Units (“NF UNITS”) equivalent to 50 mcg of active coenzyme cobalamin (as cobamamide concentrate with intrinsic factor) ALSO CONTAINS: 1 Folinic acid (B9-vitamer) 2.5 mg + 1 Control-release, citrated folic acid, DHF (B9-Provitamin) 1 mg 2 Levomefolic acid (B9 & B12- cofactor) 5.23 mg 1 6 mg DFE folate (vitamin B9) 2 9 mg DFE l-methylfolate magnesium (molar equivalent). FUNCTIONAL EXCIPIENTS: 13.6 mg FeGC as ferrous glycine cysteinate (1.5 mg 3 3,4 elemental iron ) [colorant], 25 mg ascorbates (24 mg magnesium l-ascorbate, 1 mg zinc l-ascorbate) [antioxidant], at least 23.33 mg phospholipid-omega3 complex5 [marine lipids], 500 mcg betaine (trimethylglycine) [acidifier], 1 mg magnesium l-threonate [stabilizer]. -

Actin Gesting That Cytoskeletal Laments May Be Exploited to Supplement Chemotherapeutic Approaches Currently Used Microfilaments in the Clinical Setting
Biochimica et Biophysica Acta 1846 (2014) 599–616 Contents lists available at ScienceDirect Biochimica et Biophysica Acta journal homepage: www.elsevier.com/locate/bbacan Review Exploiting the cytoskeletal filaments of neoplastic cells to potentiate a novel therapeutic approach Matthew Trendowski ⁎ Department of Biology, Syracuse University, 107 College Place, Syracuse, NY 13244, USA article info abstract Article history: Although cytoskeletal-directed agents have been a mainstay in chemotherapeutic protocols due to their ability to Received 2 August 2014 readily interfere with the rapid mitotic progression of neoplastic cells, they are all microtubule-based drugs, and Received in revised form 19 September 2014 there has yet to be any microfilament- or intermediate filament-directed agents approved for clinical use. There Accepted 21 September 2014 are many inherent differences between the cytoskeletal networks of malignant and normal cells, providing an Available online 5 October 2014 ideal target to attain preferential damage. Further, numerous microfilament-directed agents, and an intermediate fi fi Keywords: lament-directed agent of particular interest (withaferin A) have demonstrated in vitro and in vivo ef cacy, sug- fi Actin gesting that cytoskeletal laments may be exploited to supplement chemotherapeutic approaches currently used Microfilaments in the clinical setting. Therefore, this review is intended to expose academics and clinicians to the tremendous Intermediate filaments variety of cytoskeletal filament-directed agents that are currently available for further chemotherapeutic evalu- Drug synergy ation. The mechanisms by which microfilament directed- and intermediate filament-directed agents damage Chemotherapy malignant cells are discussed in detail in order to establish how the drugs can be used in combination with each other, or with currently approved chemotherapeutic agents to generate a substantial synergistic attack, potentially establishing a new paradigm of chemotherapeutic agents. -

Ribozymes Targeted to the Mitochondria Using the 5S Ribosomal Rna
RIBOZYMES TARGETED TO THE MITOCHONDRIA USING THE 5S RIBOSOMAL RNA By JENNIFER ANN BONGORNO A DISSERTATION PRESENTED TO THE GRADUATE SCHOOL OF THE UNIVERSITY OF FLORIDA IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY UNIVERSITY OF FLORIDA 2005 Copyright 2005 by Jennifer Bongorno To my grandmother, Hazel Traster Miller, whose interest in genealogy sparked my interest in genetics, and without whose mitochondria I would not be here ACKNOWLEDGMENTS I would like to thank all the members of the Lewin lab; especially my mentor, Al Lewin. Al was always there for me with suggestions and keeping me motivated. He and the other members of the lab were like my second family; I would not have had an enjoyable experience without them. Diana Levinson and Elizabeth Bongorno worked with me on the fourth and third mouse transfections respectively. Joe Hartwich and Al Lewin tested some of the ribozymes in vitro and cloned some of the constructs I used. James Thomas also helped with cloning and was an invaluable lab manager. Verline Justilien worked on a related project and was a productive person with whom to bounce ideas back and forth. Lourdes Andino taught me how to use the new phosphorimager for my SYBR Green-stained gels. Alan White was there through it all, like the older brother I never had. Mary Ann Checkley was with me even longer than Alan, since we both came to Florida from Ohio Wesleyan, although she did manage to graduate before me. Jia Liu and Frederic Manfredsson were there when I needed a beer. -

Into the Mitochondrial and Nuclear DNA and RNA of Two Transplantable Hepatomas (3924A and H-35Tc2) and Host Livers'
[CANCERRESEARCH28.2164-2167,October1968] Brief Communication Comparative Incorporation of Tritiated Thymidine and Cytidine into the Mitochondrial and Nuclear DNA and RNA of Two Transplantable Hepatomas (3924A and H-35tc2) and Host Livers' Lillie 0. Chang, Harold P.Morris,2and William B. Looney Radiobiology and Biophysics Laboratory, Department of Radiology, University of Virginia School of Medicine, Charlottesville, Vir ginia f@29O1(L. 0. C. and W. B. L.), and Laboratory of Biochemistry, National Cancer institute, NIH, Bethesda, Maryland ?LXJ114 (H. P. M.) Summary interesting questions as to the different precursor pools and It has been found that the specific activity of thymidine-5- the biosynthetic pathways of nucleic acid synthesis in the methyl-3H incorporation into the mitochondrial DNA of two mitochondria and nuclei of the hepatomas as well as of their transplantable hepatomas (3924A and H-35tc2) and host livers host livers. The purpose of this communication is to report the is less than the specific activity of the nuclear DNA. However, relative rates of uptake of the labeled thymidine and cytidine the ratio of mitochondrial DNA to nuclear DNA specific ac into DNA, and the incorporation of cytidine into RNA of the tivity in both hepatomas and host livers is 10—100times greater mitochondrial and nuclear fractions of Hepatomas 3924A and H-35tc2 and their host livers. following cytidine-5-3H administration than the ratio following thymidine-5-methyl-3H administration. This marked difference in the ratios of the specific activities of thymidine and cytidine Materials and Methods in mitochondrial and nuclear DNA of hepatomas and host livers The growth patterns of transplantable rat hepatoma lines appears to be the result of both an increase in cytidine incor have been described by Morris (8, 9) . -

Aberrant Gene Expression: Diagnostic Markers and Therapeutic Targets for Pancreatic Cancer
ABERRANT GENE EXPRESSION: DIAGNOSTIC MARKERS AND THERAPEUTIC TARGETS FOR PANCREATIC CANCER Jeran Kent Stratford A dissertation submitted to the faculty of the University of North Carolina at Chapel Hill in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Pharmacology. Chapel Hill 2014 Approved by: Jen Jen Yeh Channing J. Der Gary L. Johnson Adrienne D. Cox Timothy C. Elston © 2014 Jeran Kent Stratford ALL RIGHTS RESERVED ii ABSTRACT Jeran Kent Stratford: Aberrant gene expression: diagnostic markers and therapeutic targets for pancreatic cancer (Under the direction of Jen Jen Yeh and Channing J. Der) Pancreatic ductal adenocarcinoma (PDAC) is an aggressive cancer and the fourth leading cause of cancer-related death in the United States. The overall median survival for patients diagnosed with PDAC is five to eight months. The poor outcome is due, in part, to a lack of disease-specific symptoms that can be used for early detection, and as such, most patients present with locally advanced or metastatic disease at the time of diagnosis. Therefore, the need for diagnostic tools is both great and urgent. Furthermore, current chemotherapies have low response rates and high toxicity, limiting their use, and there are currently no effective targeted therapies for PDAC. Therefore, a greater understanding of the underlying biology of pancreatic cancer is needed to identify tumor-specific vulnerabilities that can be therapeutically exploited. Pancreatic cancer development is driven by genomic changes that alter gene expression. Aberrant gene expression produces changes in protein expression, which in turn may confer growth advantages to the tumor; often the tumor then develops a dependency on continued aberrant gene and protein expression.