The Role of Starch in the Day/Night Re-Programming of Stomata in Plants with Crassulacean Acid Metabolism Natalia Hurtado Castan
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

FLOWERS Herbaceous Perennials No
G A R D E N I N G S E R I E S FLOWERS Herbaceous Perennials no. 7.405 by M. Meehan, J.E. Klett and R.A. Cox 1 An ever-expanding palette of perennials lets home gardeners create showy collections of herbaceous perennials. Quick Facts... Under normal growing conditions, perennials live many years, dying back to the ground each winter. They quickly establish These herbaceous perennials themselves in a few growing seasons and create a are best adapted for Colorado’s backbone for the flower garden. lower elevations. Plants vary in flower color, bloom time, height, foliage texture and environmental Herbaceous perennials differ requirements. Environmental requirements include in bloom period, flower color, sun exposure, soil conditions and water needs. height, foliage texture and The key to a successful perennial garden is to environmental requirements. choose plants whose requirements match your site’s conditions. Environmental requirements Table 1 lists perennials adapted to the broad include sun and wind exposure range of growing conditions in Colorado’s lower or lack of it, soil conditions and elevations. Many also do well at higher elevations, but water needs. for a more specific listing of higher elevation perennials, see fact sheet 7.406, Flowers for Mountain Communities. Matching the perennial plant to More information on design and maintenance of perennial the site conditions produces a gardens can be found in 7.402, Perennial Gardening. Also see 7.840, Vegetable Garden: Soil Management and Fertilization. successful perennial garden. Key to Table 1: a only most common cultivars are listed. b Not Important. -

Bright Spots Plant Material Location
Bright Spots A SELF GUIDED WALKING TOUR September 13, 2017 Did you know…? Usually when we think of azaleas we think of Spring, but there are some beauties abloom in the garden right now. These are reblooming azaleas. They have already produced Spring flowers for Virginia’s Garden Week, and now they are contributing to Fall color. Many of these plants are patented under the name Encore® azalea. They first became available in the late 1990s, the work of Robert E. “Buddy” Lee, a plant breeder and nurseryman from Louisiana. This group derives from a cross between spring blooming azaleas and Rhododendron oldhamii “Fourth of July”. The garden’s Plant Explorer database lists 22 varieties of Encore® azaleas. Many have the word “Autumn” in their cultivar name. Several are in bloom now, including ‘Autumn Royalty’ and ‘Autumn Sundance’ listed below. Not much published material is available on these rebloomers, but the article by Will Ferrell (not the actor) in The Azalean (below) is a knowledgeable evaluation of the merits of many specific cultivars. Besides the Encore® series there are at least two other patented reblooming lines: ReBLOOM™ Azaleas by breeder Bob Head and Bloom-a-thon from Monrovia. To sow confusion, I must note that the Azalea ‘August to Frost’ with its bright white blossoms (along the Flagler walk) is not considered a rebloomer. It was hybridized by M. B. Matlack in 1940, possibly a cross between R. mucronatum var. mucronatum and an unknown species. Sources: http://www.encoreazalea.com/; “Personal Observations on Encore® Azaleas in a Zone 7 Garden”, Will Ferrell, The Azalean, Fall 2013, pp. -
Abbotsford Water Efficient Plant List Minimum Soil Depth Botanical Name Common Name Type (Mm)
Abbotsford Water Efficient Plant List Minimum Soil Depth Botanical name Common name type (mm) ANNUALS Argemone grandifolia Statice/Sea Lavendar A Begonia x hybrida 'Dragon Wings' Dragon Wing begonia A 150 Bracteantha species Strawflower Calendula officinalis Calendula A 150 Coleus ssp. Coleus A 150 Cosmos bipinnatus Garden cosmos A 150 Cuphea llavea Cuphea A 150 Dyssocua tenuiloba Dahlberg Daisey A Eschscholzia californica California poppy A 150 Gazania spendens Gazania A Lantana camara Lantana A 150 Lobularia maritima Sweet alyssum A 150 Nigella damascena Love-in-the-mist A 150 Oesteopermum African daisy A 150 Pelargonium species Geranium A Portulaca oleracea Portulaca A Salvia guaranitica Anise-scented sage A 150 Scaevola aemula Fan flower A 150 Targetes erecta Marogold A Targetes erecta Marogold A Viola x wittrockiana Pansy A 150 Zinnia ssp. Zinnia A 150 Verbascum bombyciferum 'Arctic Summer' Broussa mullein A 150 Verbascum phlomoides 'Spica' Orange mullein A 150 BULBS Allium christophii Star of Persia B 150 Allium karataviense Turkestan onion B 150 Chionodoxa forbesii 'Pink Giant' Glory of the snow B 150 Colchicum autumnale Autumn crocus B 150 Crocus ssp. Crocus B 150 Eranthis hyemalis Winter aconite B 150 Erythronium ssp. Fawn lily B 150 Eucomis Pineapple lily B 150 Fritillaria meleagris Checkered lily B 150 Galanthus ssp. Snowdrop B 150 Iris reticulata Dwarf iris B 150 Muscari ssp. Grape hyacinth B 150 Narcissus Daffodil B 150 Nerine bowdenii Bowden lily B 150 Ornithogalum nutans Drooping Star of Bethlehem/Silverbells B 150 Scilla -

CRASSULACEAE 景天科 Jing Tian Ke Fu Kunjun (傅坤俊 Fu Kun-Tsun)1; Hideaki Ohba 2 Herbs, Subshrubs, Or Shrubs
Flora of China 8: 202–268. 2001. CRASSULACEAE 景天科 jing tian ke Fu Kunjun (傅坤俊 Fu Kun-tsun)1; Hideaki Ohba 2 Herbs, subshrubs, or shrubs. Stems mostly fleshy. Leaves alternate, opposite, or verticillate, usually simple; stipules absent; leaf blade entire or slightly incised, rarely lobed or imparipinnate. Inflorescences terminal or axillary, cymose, corymbiform, spiculate, racemose, paniculate, or sometimes reduced to a solitary flower. Flowers usually bisexual, sometimes unisexual in Rhodiola (when plants dioecious or rarely gynodioecious), actinomorphic, (3 or)4– 6(–30)-merous. Sepals almost free or basally connate, persistent. Petals free or connate. Stamens as many as petals in 1 series or 2 × as many in 2 series. Nectar scales at or near base of carpels. Follicles sometimes fewer than sepals, free or basally connate, erect or spreading, membranous or leathery, 1- to many seeded. Seeds small; endosperm scanty or not developed. About 35 genera and over 1500 species: Africa, America, Asia, Europe; 13 genera (two endemic, one introduced) and 233 species (129 endemic, one introduced) in China. Some species of Crassulaceae are cultivated as ornamentals and/or used medicinally. Fu Shu-hsia & Fu Kun-tsun. 1984. Crassulaceae. In: Fu Shu-hsia & Fu Kun-tsun, eds., Fl. Reipubl. Popularis Sin. 34(1): 31–220. 1a. Stamens in 1 series, usually as many as petals; flowers always bisexual. 2a. Leaves always opposite, joined to form a basal sheath; inflorescences axillary, often shorter than subtending leaf; plants not developing enlarged rootstock ................................................................ 1. Tillaea 2b. Leaves alternate, occasionally opposite proximally; inflorescence terminal, often very large; plants sometimes developing enlarged, perennial rootstock. -

В Крыму Family Crassulaceae St.-Hil. in the Crimea
Turczaninowia 23, 3: 158–184 (2020) ISSN 1560–7259 (print edition) DOI: 10.14258/turczaninowia.23.3.15 TURCZANINOWIA http://turczaninowia.asu.ru ISSN 1560–7267 (online edition) УДК 582.715:581.9(477.75) Семейство толстянковые (Crassulaceae St.-Hil.) в Крыму В. В. Бялт1, 2 1 Ботанический институт им. В. Л. Комарова РАН, ул. Проф. Попова, д. 2, г. Санкт-Петербург, 197376, Россия. E-mails: [email protected], [email protected] 2 Санкт-Петербургский государственный лесотехнический университет, Институтский пер., д. 5, г. Санкт-Петербург, 194021, Россия Ключевые слова: география растений, Крымский полуостров, новая комбинация, систематика, толстянковые, флора, Crassulaceae. Аннотация. В статье дается критический обзор сем. Crassulaceae St.-Hil. (толстянковые) для Крыма, вклю- чающий в себя информацию о 19 дикорастущих и дичающих, а также о 19 более-менее широко культиви- руемых в открытом грунте видах. Предложены две новые комбинации: Crassulaceae subtrib. Sempervivinae (A. Berger) Byalt, comb. et stat. nov. и Sedum subgen. Petrosedum (Grulich) Byalt, comb. et stat. nov. Обра- зец И. Аммана, хранящийся в Гербарии БИН РАН (LE), выбран в качестве лектотипа для Sedum hybridum L. (= Aizopsis hybrida (L.) Grulich). Также предложены лектотипы для названий Crassula caespitosa Cav. (= Sedum caespitosum (cav.) DC.) и Sedum sexfidum Fisch. Мы приводим конспект и ключи для определения родов и видов дикорастущих толстянковых, а также отдельно конспект и ключи для основных культивируемых в открытом грунте видов Crassulaceae. В конспекте для дикорастущих видов использована система H. ‘t Hart (1995) с неко- торыми изменениями. Для некоторых дикорастущих и дичающих видов толстянковых приведены картосхемы их распространения на полуострове. Статья иллюстрирована 10 фотографиями. Family Crassulaceae St.-Hil. in the Crimea V. -

PERENNIAL PLANTS Plant Name Common Name Height Colour Bl Time Special Conditions Country S
PERENNIAL PLANTS Plant Name Common Name Height Colour Bl Time Special Conditions Country S. Europe, NW Acanthus mollis Bear's Breeches to 5' (1.5m) white fls. with purple shaded bracts l summer z7 sun/pt.shade,well drained, moist good soil Africa Acanthus spinosus Bear's Breeches to 5' (150cm) white flowers with purple bracts lsp-msum z5 sun/pt.shade, good soil, tolerates dry heat Italy to W Turkey Aconitum Monkshood large dark blue flowers l summer z5 sun/part shade, cool moist fertile soil Aconitum Monkshood dark blue flowers l summer z5 sun/part shade, cool moist fertile soil Monkshood (all parts are Aconitum carmichaelii to 6' (190cm) violet or blue flowers l sp to fall z3 sun/pt.shade, cool, moist, fertile soil Russia poisonous) Monkshood (all parts are Aconitum carmichaelii 'Barker's Variety' to 6' (190cm) deep violet flowers early fall z3 sun/pt.shade, cool, moist, fertile soil poisonous) Aconitum 'Ivorine' (syn.A.septentrionale Monkshpood (all parts are to 36" (90cm) ivory flowers l spring z5 sun/pt.shade, cool, moist, fertile soil garden origin 'Ivorine') poisonous) Aconitum lycoctonum ssp.vulparia Monkshood (all parts are to 5' (1.5m) pale yellow flowered form sum/e fall z4 sun/pt.shade, cool, moist, fertile soil Europe (A.orientale of gardens) poisonous) Acorus gramineus 'Variegatus' Variegated Japanese Rush to 10" (25 cm) creamy white and green striped leaves summer z5 full sun, wet or very moist soil E Asia z4 shade/pt.sh.moist mod-fertile soil.Survives under Actaea erythrocarpa (syn. A.spicata var. rubra) 24" (60cm) racemes of white flowers,red berries late spring Euro. -

Tracheophyte of Xiao Hinggan Ling in China: an Updated Checklist
Biodiversity Data Journal 7: e32306 doi: 10.3897/BDJ.7.e32306 Taxonomic Paper Tracheophyte of Xiao Hinggan Ling in China: an updated checklist Hongfeng Wang‡§, Xueyun Dong , Yi Liu|,¶, Keping Ma | ‡ School of Forestry, Northeast Forestry University, Harbin, China § School of Food Engineering Harbin University, Harbin, China | State Key Laboratory of Vegetation and Environmental Change, Institute of Botany, Chinese Academy of Sciences, Beijing, China ¶ University of Chinese Academy of Sciences, Beijing, China Corresponding author: Hongfeng Wang ([email protected]) Academic editor: Daniele Cicuzza Received: 10 Dec 2018 | Accepted: 03 Mar 2019 | Published: 27 Mar 2019 Citation: Wang H, Dong X, Liu Y, Ma K (2019) Tracheophyte of Xiao Hinggan Ling in China: an updated checklist. Biodiversity Data Journal 7: e32306. https://doi.org/10.3897/BDJ.7.e32306 Abstract Background This paper presents an updated list of tracheophytes of Xiao Hinggan Ling. The list includes 124 families, 503 genera and 1640 species (Containing subspecific units), of which 569 species (Containing subspecific units), 56 genera and 6 families represent first published records for Xiao Hinggan Ling. The aim of the present study is to document an updated checklist by reviewing the existing literature, browsing the website of National Specimen Information Infrastructure and additional data obtained in our research over the past ten years. This paper presents an updated list of tracheophytes of Xiao Hinggan Ling. The list includes 124 families, 503 genera and 1640 species (Containing subspecific units), of which 569 species (Containing subspecific units), 56 genera and 6 families represent first published records for Xiao Hinggan Ling. The aim of the present study is to document an updated checklist by reviewing the existing literature, browsing the website of National Specimen Information Infrastructure and additional data obtained in our research over the past ten years. -

Information to Users
INFORMATION TO USERS This manuscript has been reproduced from the microfilm master UMI films the text directly from the original or copy submitted. Thus, some thesis and dissertation copies are in typewriter face, while others may be from any type of computer printer. The quality of this reproduction is dependent upon the quality of the copy submitted. Broken or indistinct print, colored or poor quality illustrations and photographs, print bleedthrough, substandard margins, and improper alignment can adversely affect reproduction. In the unlikely event that the author did not send UMI a complete manuscript and there are missing pages, these will be noted. Also, if unauthorized copyright material had to be removed, a note will indicate the deletion. Oversize materials (e.g., maps, drawings, charts) are reproduced by sectioning the original, beginning at the upper left-hand comer and continuing from left to right in equal sections with small overlaps. Photographs included in the original manuscript have been reproduced xerographically in this copy. Higher quality 6" x 9" black and white photographic prints are available for any photographs or illustrations appearing in this copy for an additional charge. Contact UMI directly to order. ProQuest Information and Learning 300 North Zeeb Road, Ann Arbor, Ml 48106-1346 USA 800-521-0600 Reproduced with permission of the copyright owner. Further reproduction prohibited without permission. Reproduced with permission of the copyright owner. Further reproduction prohibited without permission. SODIUM CHLORIDE TOLERANCE OF SELECTED HERBACEOUS PERENNIALS A N D THE EFFECTS OF SODIUM CHLORIDE O N OSM OTIC ADJUSTMENT A N D IO NIC UPTAKE IN THREE SPECIES OF HERBACEOUS PERENNIALS DISSERTATION Presented in Partial Fulfillment of the Requirements of the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Laura M. -
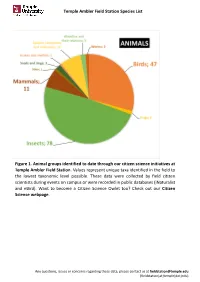
Temple Ambler Field Station Species List Figure 1. Animal Groups Identified to Date Through Our Citizen Science Initiatives at T
Temple Ambler Field Station Species List Figure 1. Animal groups identified to date through our citizen science initiatives at Temple Ambler Field Station. Values represent unique taxa identified in the field to the lowest taxonomic level possible. These data were collected by field citizen scientists during events on campus or were recorded in public databases (iNaturalist and eBird). Want to become a Citizen Science Owlet too? Check out our Citizen Science webpage. Any questions, issues or concerns regarding these data, please contact us at [email protected] (fieldstation[at}temple[dot]edu) Temple Ambler Field Station Species List Figure 2. Plant diversity identified to date in the natural environments and designed gardens of the Temple Ambler Field Station and Ambler Arboretum. These values represent unique taxa identified to the lowest taxonomic level possible. Highlighted are 14 of the 116 flowering plant families present that include 524 taxonomic groups. A full list can be found in our species database. Cultivated specimens in our Greenhouse were not included here. Any questions, issues or concerns regarding these data, please contact us at [email protected] (fieldstation[at}temple[dot]edu) Temple Ambler Field Station Species List database_title Temple Ambler Field Station Species List last_update 22October2020 description This database includes all species identified to their lowest taxonomic level possible in the natural environments and designed gardens on the Temple Ambler campus. These are occurrence records and each taxon is only entered once. This is an occurrence record, not an abundance record. IDs were performed by senior scientists and specialists, as well as citizen scientists visiting campus. -

Hylotelephium Spectabile, a New Host for Carnation Tortrix Moth (Cacoecimorpha Pronubana) and Molecular Characterization in Greece
insects Communication Hylotelephium spectabile, a New Host for Carnation Tortrix Moth (Cacoecimorpha pronubana) and Molecular Characterization in Greece Konstantinos B. Simoglou 1,* , Dimitrios N. Avtzis 2, Joaquín Baixeras 3, Ioanna Sarigkoli 1 and Emmanouil Roditakis 4,* 1 Department of Quality and Phytosanitary Inspections, Rural Economy & Veterinary Directorate of Drama, 66133 Drama, Greece; [email protected] 2 Forest Research Institute, Hellenic Agricultural Organization Demeter, Vassilika, 57006 Thessaloniki, Greece; [email protected] 3 Institut Cavanilles de Biodiversitat i Biologia Evolutiva, Universitat de València, Carrer Catedràtic José, 46980 Paterna, Spain; [email protected] 4 Department of Agriculture, School of Agricultural Sciences, Hellenic Mediterranean University, 71410 Heraklion, Greece * Correspondence: [email protected] (K.B.S.); [email protected] (E.R.) Simple Summary: Hylotelephium spectabile (Saxifragales, Crassulaceae), a widespread ornamental plant was found infested by larvae of Cacoecimorpha pronubana (Lepidoptera, Tortricidae) a highly polyphagous pest of a wide range of crop, as well as ornamental plants. To the best of our knowledge, this finding suggests that H. spectabile should be considered a new host plant for C. pronubana. Moreover, molecular characterization of the Greek pest population place it along with European species clade. Citation: Simoglou, K.B.; Avtzis, D.N.; Baixeras, J.; Sarigkoli, I.; Abstract: Cacoecimorpha pronubana (Hübner) (Lepidoptera, Tortricidae) is a highly polyphagous Roditakis, E. Hylotelephium spectabile, pest of a wide range of crop and ornamental plants. It is of Mediterranean origin and widespread a New Host for Carnation Tortrix Moth (Cacoecimorpha pronubana) and in European and Mediterranean Plant Protection Organization (EPPO) region. For the first time, Molecular Characterization in Greece. -

Cutin:Cutin-Acid Endo-Transacylase (CCT), a Cuticle- Remodelling Enzyme Activity in the Plant Epidermis
Biochemical Journal (2021) 478 777–798 https://doi.org/10.1042/BCJ20200835 Research Article Cutin:cutin-acid endo-transacylase (CCT), a cuticle- remodelling enzyme activity in the plant epidermis Anzhou Xin1,*, Yue Fei2, Attila Molnar2 and Stephen C. Fry1 1The Edinburgh Cell Wall Group, Institute of Molecular Plant Sciences, The University of Edinburgh, Edinburgh EH9 3BF, U.K.; 2Institute of Molecular Plant Sciences, The University of Edinburgh, Edinburgh EH9 3BF, U.K. Correspondence: Stephen C. Fry ([email protected]) Downloaded from http://portlandpress.com/biochemj/article-pdf/478/4/777/904720/bcj-2020-0835.pdf by guest on 06 March 2021 Cutin is a polyester matrix mainly composed of hydroxy-fatty acids that occurs in the cuticles of shoots and root-caps. The cuticle, of which cutin is a major component, pro- tects the plant from biotic and abiotic stresses, and cutin has been postulated to con- strain organ expansion. We propose that, to allow cutin restructuring, ester bonds in this net-like polymer can be transiently cleaved and then re-formed (transacylation). Here, using pea epicotyl epidermis as the main model, we first detected a cutin:cutin-fatty acid endo-transacylase (CCT) activity. In-situ assays used endogenous cutin as the donor substrate for endogenous enzymes; the exogenous acceptor substrate was a radiola- belled monomeric cutin-acid, 16-hydroxy-[3H]hexadecanoic acid (HHA). High-molecular- weight cutin became ester-bonded to intact [3H]HHA molecules, which thereby became unextractable except by ester-hydrolysing alkalis. In-situ CCT activity correlated with growth rate in Hylotelephium leaves and tomato fruits, suggesting a role in loosening the outer epidermal wall during organ growth. -

Wonderful Plants Index of Names
Wonderful Plants Jan Scholten Index of names Wonderful Plants, Index of names; Jan Scholten; © 2013, J. C. Scholten, Utrecht page 1 A’bbass 663.25.07 Adansonia baobab 655.34.10 Aki 655.44.12 Ambrosia artemisiifolia 666.44.15 Aalkruid 665.55.01 Adansonia digitata 655.34.10 Akker winde 665.76.06 Ambrosie a feuilles d’artemis 666.44.15 Aambeinwortel 665.54.12 Adder’s tongue 433.71.16 Akkerwortel 631.11.01 America swamp sassafras 622.44.10 Aardappel 665.72.02 Adder’s-tongue 633.64.14 Alarconia helenioides 666.44.07 American aloe 633.55.09 Aardbei 644.61.16 Adenandra uniflora 655.41.02 Albizia julibrissin 644.53.08 American ash 665.46.12 Aardpeer 666.44.11 Adenium obesum 665.26.06 Albuca setosa 633.53.13 American aspen 644.35.10 Aardveil 665.55.05 Adiantum capillus-veneris 444.50.13 Alcea rosea 655.33.09 American century 665.23.13 Aarons rod 665.54.04 Adimbu 665.76.16 Alchemilla arvensis 644.61.07 American false pennyroyal 665.55.20 Abécédaire 633.55.09 Adlumia fungosa 642.15.13 Alchemilla vulgaris 644.61.07 American ginseng 666.55.11 Abelia longifolia 666.62.07 Adonis aestivalis 642.13.16 Alchornea cordifolia 644.34.14 American greek valerian 664.23.13 Abelmoschus 655.33.01 Adonis vernalis 642.13.16 Alecterolophus major 665.57.06 American hedge mustard 663.53.13 Abelmoschus esculentus 655.33.01 Adoxa moschatellina 666.61.06 Alehoof 665.55.05 American hop-hornbeam 644.41.05 Abelmoschus moschatus 655.33.01 Adoxaceae 666.61 Aleppo scammony 665.76.04 American ivy 643.16.05 Abies balsamea 555.14.11 Adulsa 665.62.04 Aletris farinosa 633.26.14 American