Estimation of Phylogeny of Nineteen Sedoideae Species Cultivated in Korea Inferred from Chloroplast DNA Analysis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
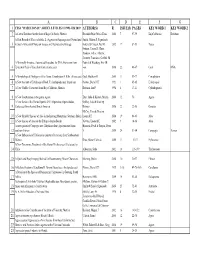
Haseltonia Articles and Authors.Xlsx
ABCDEFG 1 CSSA "HASELTONIA" ARTICLE TITLES #1 1993–#26 2019 AUTHOR(S) R ISSUE(S) PAGES KEY WORD 1 KEY WORD 2 2 A Cactus Database for the State of Baja California, Mexico Resendiz Ruiz, María Elena 2000 7 97-99 BajaCalifornia Database A First Record of Yucca aloifolia L. (Agavaceae/Asparagaceae) Naturalized Smith, Gideon F, Figueiredo, 3 in South Africa with Notes on its uses and Reproductive Biology Estrela & Crouch, Neil R 2012 17 87-93 Yucca Fotinos, Tonya D, Clase, Teodoro, Veloz, Alberto, Jimenez, Francisco, Griffith, M A Minimally Invasive, Automated Procedure for DNA Extraction from Patrick & Wettberg, Eric JB 4 Epidermal Peels of Succulent Cacti (Cactaceae) von 2016 22 46-47 Cacti DNA 5 A Morphological Phylogeny of the Genus Conophytum N.E.Br. (Aizoaceae) Opel, Matthew R 2005 11 53-77 Conophytum 6 A New Account of Echidnopsis Hook. F. (Asclepiadaceae: Stapeliae) Plowes, Darrel CH 1993 1 65-85 Echidnopsis 7 A New Cholla (Cactaceae) from Baja California, Mexico Rebman, Jon P 1998 6 17-21 Cylindropuntia 8 A New Combination in the genus Agave Etter, Julia & Kristen, Martin 2006 12 70 Agave A New Series of the Genus Opuntia Mill. (Opuntieae, Opuntioideae, Oakley, Luis & Kiesling, 9 Cactaceae) from Austral South America Roberto 2016 22 22-30 Opuntia McCoy, Tom & Newton, 10 A New Shrubby Species of Aloe in the Imatong Mountains, Southern Sudan Leonard E 2014 19 64-65 Aloe 11 A New Species of Aloe on the Ethiopia-Sudan Border Newton, Leonard E 2002 9 14-16 Aloe A new species of Ceropegia sect. -

A Numerical Taxonomy of the Genus Rosularia (Dc.) Stapf from Pakistan and Kashmir
Pak. J. Bot., 44(1): 349-354, 2012. A NUMERICAL TAXONOMY OF THE GENUS ROSULARIA (DC.) STAPF FROM PAKISTAN AND KASHMIR GHULAM RASOOL SARWAR* AND MUHAMMAD QAISER Centre for Plant Conservation, University of Karachi, Karachi-75270, Pakistan Federal Urdu University of Arts, Science and Technology, Gulshan-e-Iqbal, Karachi, Pakistan Abstract Numerical analysis of the taxa belonging to the genus Rosularia (DC.) Stapf was carried out to find out their phenetic relationship. Data from different disciplines viz. general, pollen and seed morphology, chemistry and distribution pattern were used. As a result of cluster analysis two distinct groups are formed. Out of which one group consists of R. sedoides (Decne.) H. Ohba and R. alpestris A. Boriss. while other group comprises R. adenotricha (Wall. ex Edgew.) Jansson ssp. adenotricha , R. adenotricha ssp. chitralica, G.R. Sarwar, R. rosulata (Edgew.) H. Ohba and R. viguieri (Raym.-Hamet ex Frod.) G.R. Sarwar. Distribution maps of all the taxa, along with key to the taxa are also presented. Introduction studied the genus Rosularia and indicated that the genus is polyphyletic. Mayuzumi & Ohba (2004) analyzed the Rosularia is a small genus composed of 28 species, relationships within the genus Rosularia. According to distributed in arid or semiarid regions ranging from N. different workers Rosularia is polyphyletic. Africa to C. Asia through E. Mediterranean (Mabberley, There are no reports on numerical studies of 2008). Some of the taxa of Rosularia are in general Crassulaceae except the genus Sedum from Pakistan cultivation and several have great appeal due to their (Sarwar & Qaiser, 2011). The primary aim of this study is extraordinarily regular rosettes on the leaf colouring in to analyze diagnostic value of morphological characters in various seasons. -

Hylotelephium National Protocol: NP/HYL/1.Rev ______Botanical Taxon: Hylotelephium Telephium (L.) H
Hylotelephium National protocol: NP/HYL/1.rev ____________________________________________________________________ Botanical taxon: Hylotelephium telephium (L.) H. Ohba x H. spectabile (Boreau) H. Ohba (syn. Sedum telephium L. x S. spectabile Boreau) Common Name (when known): Sedum Date of preparation of NP: 28-08-2012 Date of revision of NP: 01-01-2020 NP revised by: W.A. Wietsma Sample to be examined: VEGETATIVE Number of growing cycles: 1 year Closing date for applications: 1/12 Submission date/period: 1/4 - 30/4 Seed/Plant Quantity: 24 young plants able to show all their characteristics during the first year of examination Special conditions sample: None Test station address: Naktuinbouw, Sotaweg 22, 2371 AA, Roelofarendsveen Name: Team Support Variety Testing Department E-mail: [email protected] List of grouping characteristics: NO, (if yes put as annex) Minimum number of plants in trial: Vegetative: 20 Seed: not appl. Minimum number of plants observed by measuring or counting: Vegetative: 1 Seed: not appl. Give description of when observations on the flower should take place: At full flowering Give description of when/where observations on the leaf should take place: At full flowering Give description of when/where the other observations should take place: At full flowering Test will take place: OUTDOOR Uniformity: A population standard of 1% with an acceptance probability of at least 95%. Number of Off-types allowed: one off-type allowed in a sample size of 24 Table of characteristics: PRESENT (see annex) (if present, please annex the table of characteristics and explanations) Literature: PRESENT (when present, please annex to this document) Page 1 of 3 Table of characteristics Hylotelephium 1. -

The Vascular Plants of Massachusetts
The Vascular Plants of Massachusetts: The Vascular Plants of Massachusetts: A County Checklist • First Revision Melissa Dow Cullina, Bryan Connolly, Bruce Sorrie and Paul Somers Somers Bruce Sorrie and Paul Connolly, Bryan Cullina, Melissa Dow Revision • First A County Checklist Plants of Massachusetts: Vascular The A County Checklist First Revision Melissa Dow Cullina, Bryan Connolly, Bruce Sorrie and Paul Somers Massachusetts Natural Heritage & Endangered Species Program Massachusetts Division of Fisheries and Wildlife Natural Heritage & Endangered Species Program The Natural Heritage & Endangered Species Program (NHESP), part of the Massachusetts Division of Fisheries and Wildlife, is one of the programs forming the Natural Heritage network. NHESP is responsible for the conservation and protection of hundreds of species that are not hunted, fished, trapped, or commercially harvested in the state. The Program's highest priority is protecting the 176 species of vertebrate and invertebrate animals and 259 species of native plants that are officially listed as Endangered, Threatened or of Special Concern in Massachusetts. Endangered species conservation in Massachusetts depends on you! A major source of funding for the protection of rare and endangered species comes from voluntary donations on state income tax forms. Contributions go to the Natural Heritage & Endangered Species Fund, which provides a portion of the operating budget for the Natural Heritage & Endangered Species Program. NHESP protects rare species through biological inventory, -

Front Matter
Cambridge University Press 978-1-107-01276-9 - The Systematics Association Special Volume 83: Early Events in Monocot Evolution Edited by Paul Wilkin and Simon J. Mayo Frontmatter More information Early Events in Monocot Evolution Tracing the evolution of one of the most ancient major branches of flowering plants, this is a wide-ranging survey of state-of-the-art research on the early clades of the monocot phylogenetic tree. It explores a series of broad but linked themes, providing for the first time a detailed and coherent view of the taxa of the early monocot lineages, how they diversified and their importance in monocots as a whole. Featuring contributions from leaders in the field, the chapters trace the evolu- tion of the monocots from largely aquatic ancestors. Topics covered include the rapidly advancing field of monocot fossils, aquatic adaptations in pollen and anther structure and pollination strategies, and floral developmental morphology. The book also presents a new phylogenetic tree of early monocots based on sequence data from 17 plastid regions, and a review of monocot phylogeny as a whole, placing in an evolutionary context a plant group of major ecological, economic and horticultural importance. Abstracts and key words for each chapter are available for download at www. cambridge.org/9781107012769. Paul Wilkin is Lilioid and Alismatid Monocots and Ferns Team Leader in the Herbarium, Library, Art and Archives Directorate of the Royal Botanic Gardens, Kew. His main research foci are systematics of Dioscoreales (yams and their allies) and dracaenoids (dragon trees and mother-in-law’s tongues), lilioid monocots widely used in human diet and horticulture, with taxa of high conservation and ecological importance. -

FLOWERS Herbaceous Perennials No
G A R D E N I N G S E R I E S FLOWERS Herbaceous Perennials no. 7.405 by M. Meehan, J.E. Klett and R.A. Cox 1 An ever-expanding palette of perennials lets home gardeners create showy collections of herbaceous perennials. Quick Facts... Under normal growing conditions, perennials live many years, dying back to the ground each winter. They quickly establish These herbaceous perennials themselves in a few growing seasons and create a are best adapted for Colorado’s backbone for the flower garden. lower elevations. Plants vary in flower color, bloom time, height, foliage texture and environmental Herbaceous perennials differ requirements. Environmental requirements include in bloom period, flower color, sun exposure, soil conditions and water needs. height, foliage texture and The key to a successful perennial garden is to environmental requirements. choose plants whose requirements match your site’s conditions. Environmental requirements Table 1 lists perennials adapted to the broad include sun and wind exposure range of growing conditions in Colorado’s lower or lack of it, soil conditions and elevations. Many also do well at higher elevations, but water needs. for a more specific listing of higher elevation perennials, see fact sheet 7.406, Flowers for Mountain Communities. Matching the perennial plant to More information on design and maintenance of perennial the site conditions produces a gardens can be found in 7.402, Perennial Gardening. Also see 7.840, Vegetable Garden: Soil Management and Fertilization. successful perennial garden. Key to Table 1: a only most common cultivars are listed. b Not Important. -

State of New York City's Plants 2018
STATE OF NEW YORK CITY’S PLANTS 2018 Daniel Atha & Brian Boom © 2018 The New York Botanical Garden All rights reserved ISBN 978-0-89327-955-4 Center for Conservation Strategy The New York Botanical Garden 2900 Southern Boulevard Bronx, NY 10458 All photos NYBG staff Citation: Atha, D. and B. Boom. 2018. State of New York City’s Plants 2018. Center for Conservation Strategy. The New York Botanical Garden, Bronx, NY. 132 pp. STATE OF NEW YORK CITY’S PLANTS 2018 4 EXECUTIVE SUMMARY 6 INTRODUCTION 10 DOCUMENTING THE CITY’S PLANTS 10 The Flora of New York City 11 Rare Species 14 Focus on Specific Area 16 Botanical Spectacle: Summer Snow 18 CITIZEN SCIENCE 20 THREATS TO THE CITY’S PLANTS 24 NEW YORK STATE PROHIBITED AND REGULATED INVASIVE SPECIES FOUND IN NEW YORK CITY 26 LOOKING AHEAD 27 CONTRIBUTORS AND ACKNOWLEGMENTS 30 LITERATURE CITED 31 APPENDIX Checklist of the Spontaneous Vascular Plants of New York City 32 Ferns and Fern Allies 35 Gymnosperms 36 Nymphaeales and Magnoliids 37 Monocots 67 Dicots 3 EXECUTIVE SUMMARY This report, State of New York City’s Plants 2018, is the first rankings of rare, threatened, endangered, and extinct species of what is envisioned by the Center for Conservation Strategy known from New York City, and based on this compilation of The New York Botanical Garden as annual updates thirteen percent of the City’s flora is imperiled or extinct in New summarizing the status of the spontaneous plant species of the York City. five boroughs of New York City. This year’s report deals with the City’s vascular plants (ferns and fern allies, gymnosperms, We have begun the process of assessing conservation status and flowering plants), but in the future it is planned to phase in at the local level for all species. -

Classificatie Van Planten – Nieuwe Inzichten En Gevolgen Voor De Praktijk
B268_dendro_bin 03-11-2009 10:32 Pagina 4 Classificatie van planten – nieuwe inzichten en gevolgen voor de praktijk Ir. M.H.A. Hoffman Er zijn ongeveer 300.000 verschillende (hogere) plantensoorten, die onderling veel of weinig op elkaar lijken. Vroeger werden alle planten- soorten afzonderlijk benaamd, en was niet duidelijk hoe deze soorten ver- want waren. Tegenwoordig worden soorten conform het systeem van Lin- naeus ingedeeld in geslachten. Soorten die sterk verwant zijn aan elkaar hebben dezelfde geslachtsnaam. Geslachten worden op hun beurt weer ingedeeld in families en die op hun beurt weer in ordes, enzovoort. Op deze manier kent het plantenrijk een hiërarchisch indelingsysteem, met verwantschap als basis. Sterke verwantschap is meestal ook zicht- baar aan de uiterlijke gelijkenis. Soorten die veel op elkaar lijken en verwant zijn zitten in dezelfde groep.Verre verwanten zitten ver uit elkaar in het systeem. Om een plant goed te kunnen benamen, moet dus eerst uitgezocht worden aan welke andere soorten deze verwant is. Tot voor kort werd de gelijkenis van planten zen. Deze nieuwe ontwikkeling is van grote voornamelijk bepaald aan uiterlijke kenmerken invloed op de taxonomie en op het classificatie- van bijvoorbeeld bloem en blad. De afgelopen systeem van het plantenrijk. Dit heeft bijvoor- decennia kwamen daar al aanvullende criteria beeld invloed op de familie-indeling en de plaats zoals houtanatomie, pollenmorfologie, chemi- van de familie in het systeem. Maar ook op sche inhoudsstoffen en chromosoomaantallen geslachts- en soortniveau zijn er de nodige ver- bij. De afgelopen jaren hebben DNA-technie- schuivingen. Dit artikel gaat in op de ontstaans- ken een grote vlucht genomen. -

Sedum Society Newsletter(130) Pp
Open Research Online The Open University’s repository of research publications and other research outputs Kalanchoe arborescens - a Madagascan giant Journal Item How to cite: Walker, Colin (2019). Kalanchoe arborescens - a Madagascan giant. Sedum Society Newsletter(130) pp. 81–84. For guidance on citations see FAQs. c [not recorded] https://creativecommons.org/licenses/by-nc-nd/4.0/ Version: Version of Record Copyright and Moral Rights for the articles on this site are retained by the individual authors and/or other copyright owners. For more information on Open Research Online’s data policy on reuse of materials please consult the policies page. oro.open.ac.uk NUMBER 130 SEDUM SOCIETY NEWSLETTER JULY 2019 FRONT COVER Roy Mottram kindly supplied: “The Diet” copy of this Japanese herbal which is sharp and crisp (see page 97). “I counted the plates, and this copy is complete with 200 plates, in 8 parts, bound here in 2 vols. I checked for another Sedum but none are Established April 1987, now ending our present, so Maximowicz was basing his 32nd year. S. kagamontanum on this same plate, Subscriptions run from October to the following September. Anyone requesting translating the location as Mt. Kaga and to join after June, unless there is a special citing t.40 incorrectly. The "t.43" plate request, will receive his or her first number is also wrong. It is actually t.33 of Newsletter in October. If you do not the whole work, or Vol.2 t.8. The book is receive your copy by the 10th of April, July or October, or the 15th January, then bound back to front [by Western standards] please write to the editor: Ray as in all Japanese books of the day.” RM. -

Bright Spots Plant Material Location
Bright Spots A SELF GUIDED WALKING TOUR September 13, 2017 Did you know…? Usually when we think of azaleas we think of Spring, but there are some beauties abloom in the garden right now. These are reblooming azaleas. They have already produced Spring flowers for Virginia’s Garden Week, and now they are contributing to Fall color. Many of these plants are patented under the name Encore® azalea. They first became available in the late 1990s, the work of Robert E. “Buddy” Lee, a plant breeder and nurseryman from Louisiana. This group derives from a cross between spring blooming azaleas and Rhododendron oldhamii “Fourth of July”. The garden’s Plant Explorer database lists 22 varieties of Encore® azaleas. Many have the word “Autumn” in their cultivar name. Several are in bloom now, including ‘Autumn Royalty’ and ‘Autumn Sundance’ listed below. Not much published material is available on these rebloomers, but the article by Will Ferrell (not the actor) in The Azalean (below) is a knowledgeable evaluation of the merits of many specific cultivars. Besides the Encore® series there are at least two other patented reblooming lines: ReBLOOM™ Azaleas by breeder Bob Head and Bloom-a-thon from Monrovia. To sow confusion, I must note that the Azalea ‘August to Frost’ with its bright white blossoms (along the Flagler walk) is not considered a rebloomer. It was hybridized by M. B. Matlack in 1940, possibly a cross between R. mucronatum var. mucronatum and an unknown species. Sources: http://www.encoreazalea.com/; “Personal Observations on Encore® Azaleas in a Zone 7 Garden”, Will Ferrell, The Azalean, Fall 2013, pp. -

The Role of Starch in the Day/Night Re-Programming of Stomata in Plants with Crassulacean Acid Metabolism Natalia Hurtado Castan
The role of starch in the day/night re-programming of stomata in plants with Crassulacean Acid Metabolism Natalia Hurtado Castano Doctor of Philosophy School of Natural and Environmental Sciences February 2020 Declaration This thesis is submitted to Newcastle University for the degree of Doctor of Philosophy. The research detailed within was performed between the years 2015-2019 and it was supervised by Professor Anne Borland and Professor Jeremy Barnes. I certify that none of the material offered in this thesis has been previously submitted by me for a degree or any other qualification at this or any other university. Natalia Hurtado Castano ii Abstract Crassulacean acid metabolism (CAM) is a specialised type of photosynthesis characterised by the unique inverted stomatal rhythm, which increases water use efficiency (WUE) and enhances the potential for sustainable biomass production in warmer and drier conditions. Starch turnover in the mesophyll of CAM species supports nocturnal CO2 assimilation and CAM activity. In C3 plants, starch metabolism has been reported to play an important role in determining stomatal behaviour; in this case, guard cell starch degradation is triggered by blue light, producing osmolytes that promotes stomatal opening. Based on the importance of starch and the little knowledge regarding CAM stomatal behaviour, this study tested the hypothesis that starch metabolism has been re-programmed in CAM plants to enable nocturnal stomatal opening, by using biochemical and genetic characterisation of wild type and RNAi lines with curtailed starch metabolism in the constitutive CAM species Kalanchoë fedtschenkoi. Measurements of guard cell starch content over 24 hours in wild type plants of K. -

CRASSULACEAE 景天科 Jing Tian Ke Fu Kunjun (傅坤俊 Fu Kun-Tsun)1; Hideaki Ohba 2 Herbs, Subshrubs, Or Shrubs
Flora of China 8: 202–268. 2001. CRASSULACEAE 景天科 jing tian ke Fu Kunjun (傅坤俊 Fu Kun-tsun)1; Hideaki Ohba 2 Herbs, subshrubs, or shrubs. Stems mostly fleshy. Leaves alternate, opposite, or verticillate, usually simple; stipules absent; leaf blade entire or slightly incised, rarely lobed or imparipinnate. Inflorescences terminal or axillary, cymose, corymbiform, spiculate, racemose, paniculate, or sometimes reduced to a solitary flower. Flowers usually bisexual, sometimes unisexual in Rhodiola (when plants dioecious or rarely gynodioecious), actinomorphic, (3 or)4– 6(–30)-merous. Sepals almost free or basally connate, persistent. Petals free or connate. Stamens as many as petals in 1 series or 2 × as many in 2 series. Nectar scales at or near base of carpels. Follicles sometimes fewer than sepals, free or basally connate, erect or spreading, membranous or leathery, 1- to many seeded. Seeds small; endosperm scanty or not developed. About 35 genera and over 1500 species: Africa, America, Asia, Europe; 13 genera (two endemic, one introduced) and 233 species (129 endemic, one introduced) in China. Some species of Crassulaceae are cultivated as ornamentals and/or used medicinally. Fu Shu-hsia & Fu Kun-tsun. 1984. Crassulaceae. In: Fu Shu-hsia & Fu Kun-tsun, eds., Fl. Reipubl. Popularis Sin. 34(1): 31–220. 1a. Stamens in 1 series, usually as many as petals; flowers always bisexual. 2a. Leaves always opposite, joined to form a basal sheath; inflorescences axillary, often shorter than subtending leaf; plants not developing enlarged rootstock ................................................................ 1. Tillaea 2b. Leaves alternate, occasionally opposite proximally; inflorescence terminal, often very large; plants sometimes developing enlarged, perennial rootstock.