Working with Molecular Genetics Chapter 4: Genomes and Chromosomes CHAPTER 4 GENOMES and CHROMOSOMES
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Engineering Curves – I
Engineering Curves – I 1. Classification 2. Conic sections - explanation 3. Common Definition 4. Ellipse – ( six methods of construction) 5. Parabola – ( Three methods of construction) 6. Hyperbola – ( Three methods of construction ) 7. Methods of drawing Tangents & Normals ( four cases) Engineering Curves – II 1. Classification 2. Definitions 3. Involutes - (five cases) 4. Cycloid 5. Trochoids – (Superior and Inferior) 6. Epic cycloid and Hypo - cycloid 7. Spiral (Two cases) 8. Helix – on cylinder & on cone 9. Methods of drawing Tangents and Normals (Three cases) ENGINEERING CURVES Part- I {Conic Sections} ELLIPSE PARABOLA HYPERBOLA 1.Concentric Circle Method 1.Rectangle Method 1.Rectangular Hyperbola (coordinates given) 2.Rectangle Method 2 Method of Tangents ( Triangle Method) 2 Rectangular Hyperbola 3.Oblong Method (P-V diagram - Equation given) 3.Basic Locus Method 4.Arcs of Circle Method (Directrix – focus) 3.Basic Locus Method (Directrix – focus) 5.Rhombus Metho 6.Basic Locus Method Methods of Drawing (Directrix – focus) Tangents & Normals To These Curves. CONIC SECTIONS ELLIPSE, PARABOLA AND HYPERBOLA ARE CALLED CONIC SECTIONS BECAUSE THESE CURVES APPEAR ON THE SURFACE OF A CONE WHEN IT IS CUT BY SOME TYPICAL CUTTING PLANES. OBSERVE ILLUSTRATIONS GIVEN BELOW.. Ellipse Section Plane Section Plane Hyperbola Through Generators Parallel to Axis. Section Plane Parallel to end generator. COMMON DEFINATION OF ELLIPSE, PARABOLA & HYPERBOLA: These are the loci of points moving in a plane such that the ratio of it’s distances from a fixed point And a fixed line always remains constant. The Ratio is called ECCENTRICITY. (E) A) For Ellipse E<1 B) For Parabola E=1 C) For Hyperbola E>1 Refer Problem nos. -

Genome Analysis of the Smallest Free-Living Eukaryote Ostreococcus
Genome analysis of the smallest free-living eukaryote SEE COMMENTARY Ostreococcus tauri unveils many unique features Evelyne Derellea,b, Conchita Ferrazb,c, Stephane Rombautsb,d, Pierre Rouze´ b,e, Alexandra Z. Wordenf, Steven Robbensd, Fre´ de´ ric Partenskyg, Sven Degroeved,h, Sophie Echeynie´ c, Richard Cookei, Yvan Saeysd, Jan Wuytsd, Kamel Jabbarij, Chris Bowlerk, Olivier Panaudi, BenoıˆtPie´ gui, Steven G. Ballk, Jean-Philippe Ralk, Franc¸ois-Yves Bougeta, Gwenael Piganeaua, Bernard De Baetsh, Andre´ Picarda,l, Michel Delsenyi, Jacques Demaillec, Yves Van de Peerd,m, and Herve´ Moreaua,m aObservatoire Oce´anologique, Laboratoire Arago, Unite´Mixte de Recherche 7628, Centre National de la Recherche Scientifique–Universite´Pierre et Marie Curie-Paris 6, BP44, 66651 Banyuls sur Mer Cedex, France; cInstitut de Ge´ne´ tique Humaine, Unite´Propre de Recherche 1142, Centre National de la Recherche Scientifique, 141 Rue de Cardonille, 34396 Montpellier Cedex 5, France; dDepartment of Plant Systems Biology, Flanders Interuniversity Institute for Biotechnology and eLaboratoire Associe´de l’Institut National de la Recherche Agronomique (France), Ghent University, Technologiepark 927, 9052 Ghent, Belgium; fRosenstiel School of Marine and Atmospheric Science, University of Miami, 4600 Rickenbacker Causeway, Miami, FL 33149; gStation Biologique, Unite´Mixte de Recherche 7144, Centre National de la Recherche Scientifique–Universite´Pierre et Marie Curie-Paris 6, BP74, 29682 Roscoff Cedex, France; hDepartment of Applied Mathematics, Biometrics and -

The Transcriptional Landscape of a Rewritten Bacterial Genome Reveals Control Elements and Genome Design Principles ✉ ✉ Mariëlle J
ARTICLE https://doi.org/10.1038/s41467-021-23362-y OPEN The transcriptional landscape of a rewritten bacterial genome reveals control elements and genome design principles ✉ ✉ Mariëlle J. F. M. van Kooten 1 , Clio A. Scheidegger1, Matthias Christen1 & Beat Christen 1 Sequence rewriting enables low-cost genome synthesis and the design of biological systems with orthogonal genetic codes. The error-free, robust rewriting of nucleotide sequences can 1234567890():,; be achieved with a complete annotation of gene regulatory elements. Here, we compare transcription in Caulobacter crescentus to transcription from plasmid-borne segments of the synthesized genome of C. ethensis 2.0. This rewritten derivative contains an extensive amount of supposedly neutral mutations, including 123’562 synonymous codon changes. The tran- scriptional landscape refines 60 promoter annotations, exposes 18 termination elements and links extensive transcription throughout the synthesized genome to the unintentional intro- duction of sigma factor binding motifs. We reveal translational regulation for 20 CDS and uncover an essential translational regulatory element for the expression of ribosomal protein RplS. The annotation of gene regulatory elements allowed us to formulate design principles that improve design schemes for synthesized DNA, en route to a bright future of iteration- free programming of biological systems. 1 Institute of Molecular Systems Biology, Department of Biology, Eidgenössische Technische Hochschule Zürich, Zürich, Switzerland. ✉ email: [email protected]; [email protected] NATURE COMMUNICATIONS | (2021) 12:3053 | https://doi.org/10.1038/s41467-021-23362-y | www.nature.com/naturecommunications 1 ARTICLE NATURE COMMUNICATIONS | https://doi.org/10.1038/s41467-021-23362-y e can program biological systems with DNA that is In previous work, we synthesized and assembled the rewritten Wbased on native nucleotide sequences. -

Insights Into Comparative Genomics, Codon Usage Bias, And
plants Article Insights into Comparative Genomics, Codon Usage Bias, and Phylogenetic Relationship of Species from Biebersteiniaceae and Nitrariaceae Based on Complete Chloroplast Genomes Xiaofeng Chi 1,2 , Faqi Zhang 1,2 , Qi Dong 1,* and Shilong Chen 1,2,* 1 Key Laboratory of Adaptation and Evolution of Plateau Biota, Northwest Institute of Plateau Biology, Chinese Academy of Sciences, Xining 810008, China; [email protected] (X.C.); [email protected] (F.Z.) 2 Qinghai Provincial Key Laboratory of Crop Molecular Breeding, Northwest Institute of Plateau Biology, Chinese Academy of Sciences, Xining 810008, China * Correspondence: [email protected] (Q.D.); [email protected] (S.C.) Received: 29 October 2020; Accepted: 17 November 2020; Published: 18 November 2020 Abstract: Biebersteiniaceae and Nitrariaceae, two small families, were classified in Sapindales recently. Taxonomic and phylogenetic relationships within Sapindales are still poorly resolved and controversial. In current study, we compared the chloroplast genomes of five species (Biebersteinia heterostemon, Peganum harmala, Nitraria roborowskii, Nitraria sibirica, and Nitraria tangutorum) from Biebersteiniaceae and Nitrariaceae. High similarity was detected in the gene order, content and orientation of the five chloroplast genomes; 13 highly variable regions were identified among the five species. An accelerated substitution rate was found in the protein-coding genes, especially clpP. The effective number of codons (ENC), parity rule 2 (PR2), and neutrality plots together revealed that the codon usage bias is affected by mutation and selection. The phylogenetic analysis strongly supported (Nitrariaceae (Biebersteiniaceae + The Rest)) relationships in Sapindales. Our findings can provide useful information for analyzing phylogeny and molecular evolution within Biebersteiniaceae and Nitrariaceae. -

Mutation Bias Shapes Gene Evolution in Arabidopsis Thaliana
bioRxiv preprint doi: https://doi.org/10.1101/2020.06.17.156752; this version posted June 18, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Mutation bias shapes gene evolution in Arabidopsis thaliana 1,2† 1 1 3,4 Monroe, J. Grey , Srikant, Thanvi , Carbonell-Bejerano, Pablo , Exposito-Alonso, Moises , 5 6 7 1† Weng, Mao-Lun , Rutter, Matthew T. , Fenster, Charles B. , Weigel, Detlef 1 Department of Molecular Biology, Max Planck Institute for Developmental Biology, 72076 Tübingen, Germany 2 Department of Plant Sciences, University of California Davis, Davis, CA 95616, USA 3 Department of Plant Biology, Carnegie Institution for Science, Stanford, CA 94305, USA 4 Department of Biology, Stanford University, Stanford, CA 94305, USA 5 Department of Biology, Westfield State University, Westfield, MA 01086, USA 6 Department of Biology, College of Charleston, SC 29401, USA 7 Department of Biology and Microbiology, South Dakota State University, Brookings, SD 57007, USA † corresponding authors: [email protected], [email protected] Classical evolutionary theory maintains that mutation rate variation between genes should be random with respect to fitness 1–4 and evolutionary optimization of genic 3,5 mutation rates remains controversial . However, it has now become known that cytogenetic (DNA sequence + epigenomic) features influence local mutation probabilities 6 , which is predicted by more recent theory to be a prerequisite for beneficial mutation 7 rates between different classes of genes to readily evolve . To test this possibility, we used de novo mutations in Arabidopsis thaliana to create a high resolution predictive model of mutation rates as a function of cytogenetic features across the genome. -

Topography of the Histone Octamer Surface: Repeating Structural Motifs Utilized in the Docking of Nucleosomal
Proc. Natl. Acad. Sci. USA Vol. 90, pp. 10489-10493, November 1993 Biochemistry Topography of the histone octamer surface: Repeating structural motifs utilized in the docking of nucleosomal DNA (histone fold/helix-strand-helix motif/parallel fi bridge/binary DNA binding sites/nucleosome) GINA ARENTS* AND EVANGELOS N. MOUDRIANAKIS*t *Department of Biology, The Johns Hopkins University, Baltimore, MD 21218; and tDepartment of Biology, University of Athens, Athens, Greece Communicated by Christian B. Anfinsen, August 5, 1993 ABSTRACT The histone octamer core of the nucleosome is interactions. The model offers strong predictive criteria for a protein superhelix offour spirally arrayed histone dimers. The structural and genetic biology. cylindrical face of this superhelix is marked by intradimer and interdimer pseudodyad axes, which derive from the nature ofthe METHODS histone fold. The histone fold appears as the result of a tandem, parallel duplication of the "helix-strand-helix" motif. This The determination of the structure of the histone octamer at motif, by its occurrence in the four dimers, gives rise torepetitive 3.1 A has been described (3). The overall shape and volume structural elements-i.e., the "parallel 13 bridges" and the of this tripartite structure is in agreement with the results of "paired ends of helix I" motifs. A preponderance of positive three independent studies based on differing methodolo- charges on the surface of the octamer appears as a left-handed gies-i.e., x-ray diffraction, neutron diffraction, and electron spiral situated at the expected path of the DNA. We have microscopic image reconstruction (4-6). Furthermore, the matched a subset of DNA pseudodyads with the octamer identification of the histone fold, a tertiary structure motif of pseudodyads and thus have built a model of the nucleosome. -

Horizontal Gene Flow Into Geobacillus Is Constrained by the Chromosomal Organization of Growth and Sporulation
bioRxiv preprint doi: https://doi.org/10.1101/381442; this version posted August 2, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Horizontal gene flow into Geobacillus is constrained by the chromosomal organization of growth and sporulation Alexander Esin1,2, Tom Ellis3,4, Tobias Warnecke1,2* 1Molecular Systems Group, Medical Research Council London Institute of Medical Sciences, London, United Kingdom 2Institute of Clinical Sciences, Faculty of Medicine, Imperial College London, London, United Kingdom 3Imperial College Centre for Synthetic Biology, Imperial College London, London, United Kingdom 4Department of Bioengineering, Imperial College London, London, United Kingdom *corresponding author ([email protected]) 1 bioRxiv preprint doi: https://doi.org/10.1101/381442; this version posted August 2, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Abstract Horizontal gene transfer (HGT) in bacteria occurs in the context of adaptive genome architecture. As a consequence, some chromosomal neighbourhoods are likely more permissive to HGT than others. Here, we investigate the chromosomal topology of horizontal gene flow into a clade of Bacillaceae that includes Geobacillus spp. Reconstructing HGT patterns using a phylogenetic approach coupled to model-based reconciliation, we discover three large contiguous chromosomal zones of HGT enrichment. -

Section 4. Guidance Document on Horizontal Gene Transfer Between Bacteria
306 - PART 2. DOCUMENTS ON MICRO-ORGANISMS Section 4. Guidance document on horizontal gene transfer between bacteria 1. Introduction Horizontal gene transfer (HGT) 1 refers to the stable transfer of genetic material from one organism to another without reproduction. The significance of horizontal gene transfer was first recognised when evidence was found for ‘infectious heredity’ of multiple antibiotic resistance to pathogens (Watanabe, 1963). The assumed importance of HGT has changed several times (Doolittle et al., 2003) but there is general agreement now that HGT is a major, if not the dominant, force in bacterial evolution. Massive gene exchanges in completely sequenced genomes were discovered by deviant composition, anomalous phylogenetic distribution, great similarity of genes from distantly related species, and incongruent phylogenetic trees (Ochman et al., 2000; Koonin et al., 2001; Jain et al., 2002; Doolittle et al., 2003; Kurland et al., 2003; Philippe and Douady, 2003). There is also much evidence now for HGT by mobile genetic elements (MGEs) being an ongoing process that plays a primary role in the ecological adaptation of prokaryotes. Well documented is the example of the dissemination of antibiotic resistance genes by HGT that allowed bacterial populations to rapidly adapt to a strong selective pressure by agronomically and medically used antibiotics (Tschäpe, 1994; Witte, 1998; Mazel and Davies, 1999). MGEs shape bacterial genomes, promote intra-species variability and distribute genes between distantly related bacterial genera. Horizontal gene transfer (HGT) between bacteria is driven by three major processes: transformation (the uptake of free DNA), transduction (gene transfer mediated by bacteriophages) and conjugation (gene transfer by means of plasmids or conjugative and integrated elements). -

Genome Evolution: Mutation Is the Main Driver of Genome Size in Prokaryotes Gabriel A.B
Genome Evolution: Mutation Is the Main Driver of Genome Size in Prokaryotes Gabriel A.B. Marais, Bérénice Batut, Vincent Daubin To cite this version: Gabriel A.B. Marais, Bérénice Batut, Vincent Daubin. Genome Evolution: Mutation Is the Main Driver of Genome Size in Prokaryotes. Current Biology - CB, Elsevier, 2020, 30 (19), pp.R1083- R1085. 10.1016/j.cub.2020.07.093. hal-03066151 HAL Id: hal-03066151 https://hal.archives-ouvertes.fr/hal-03066151 Submitted on 15 Dec 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. DISPATCH Genome Evolution: Mutation is the Main Driver of Genome Size in Prokaryotes Gabriel A.B. Marais1, Bérénice Batut2, and Vincent Daubin1 1Université Lyon 1, CNRS, Laboratoire de Biométrie et Biologie Évolutive UMR 5558, F- 69622 Villeurbanne, France 2Albert-Ludwigs-University Freiburg, Department of Computer Science, 79110 Freiburg, Germany Summary Despite intense research on genome architecture since the 2000’s, genome-size evolution in prokaryotes has remained puzzling. Using a phylogenetic approach, a new study found that increased mutation rate is associated with gene loss and reduced genome size in prokaryotes. In 2003 [1] and later in 2007 in his book “The Origins of Genome Architecture” [2], Lynch developed his influential theory that a genome’s complexity, represented by its size, is primarily the result of genetic drift. -

Art and Engineering Inspired by Swarm Robotics
RICE UNIVERSITY Art and Engineering Inspired by Swarm Robotics by Yu Zhou A Thesis Submitted in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy Approved, Thesis Committee: Ronald Goldman, Chair Professor of Computer Science Joe Warren Professor of Computer Science Marcia O'Malley Professor of Mechanical Engineering Houston, Texas April, 2017 ABSTRACT Art and Engineering Inspired by Swarm Robotics by Yu Zhou Swarm robotics has the potential to combine the power of the hive with the sen- sibility of the individual to solve non-traditional problems in mechanical, industrial, and architectural engineering and to develop exquisite art beyond the ken of most contemporary painters, sculptors, and architects. The goal of this thesis is to apply swarm robotics to the sublime and the quotidian to achieve this synergy between art and engineering. The potential applications of collective behaviors, manipulation, and self-assembly are quite extensive. We will concentrate our research on three topics: fractals, stabil- ity analysis, and building an enhanced multi-robot simulator. Self-assembly of swarm robots into fractal shapes can be used both for artistic purposes (fractal sculptures) and in engineering applications (fractal antennas). Stability analysis studies whether distributed swarm algorithms are stable and robust either to sensing or to numerical errors, and tries to provide solutions to avoid unstable robot configurations. Our enhanced multi-robot simulator supports this research by providing real-time simula- tions with customized parameters, and can become as well a platform for educating a new generation of artists and engineers. The goal of this thesis is to use techniques inspired by swarm robotics to develop a computational framework accessible to and suitable for both artists and engineers. -

Evolution of Genome Size
Evolution of Genome Advanced article Article Contents Size • Introduction • How Much Variation Is There? Stephen I Wright, Department of Ecology and Evolutionary Biology, University • What Types of DNA Drive Genome Size of Toronto, Toronto, Ontario, Canada Variation? • Neutral Model • Nearly Neutral Model • Adaptive Hypotheses • Transposable Element Evolution • Conclusion • Acknowledgements Online posting date: 16th January 2017 The size of the genome represents one of the most in the last century. While considerable progress has been made strikingly variable yet poorly understood traits in the characterisation of the extent of genome size variation, in eukaryotic organisms. Genomic comparisons the dominant evolutionary processes driving genome size evolu- suggest that most properties of genomes tend tion remain subject to considerable debate. Large-scale genome sequencing is enabling new insights into both the proximate to increase with genome size, but the fraction causes and evolutionary forces governing genome size differ- of the genome that comprises transposable ele- ences. ments (TEs) and other repetitive elements tends to increase disproportionately. Neutral, nearly neutral and adaptive models for the evolution of How Much Variation Is There? genome size have been proposed, but strong evi- dence for the general importance of any of these Because determining the amount of DNA (deoxyribonucleic models remains lacking, and improved under- acid) in a cell has been much more straightforward and cheaper standing of factors driving the -
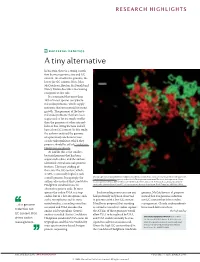
Bacterial Genetics a Tiny Alternative
RESEARCH HIGHLIGHTS BACTERIAL GENETICS A tiny alternative In bacteria, there is a strong correla- tion between genome size and GC content: the smaller the genome, the lower the GC content. Now, John McCutcheon, Bradon McDonald and Nancy Moran describe a fascinating exception to this rule. It is estimated that more than 10% of insect species carry bacte- rial endosymbionts, which supply nutrients that are essential for insect growth. The genomes of the bacte- rial endosymbionts that have been sequenced so far are much smaller than the genomes of other intracel- lular or free-living bacteria and all have a low GC content. In this study, the authors analysed the genome of a previously uncharacterized cicada endosymbiont, which they propose should be called Candidatus Hodgkinia cicadicola. At 144 kb, this is the smallest bacterial genome that has been sequenced to date, and the authors identified several unusual genomic features. The most striking of these was the GC content, which, at 58%, is unusually high for such Micrograph showing Candidatus Hodgkinia cicadicola (red) in close association with another endosymbiont, a small genome. Intriguingly, the Candidatus Sulcia muelleri (green) in the cicada Diceroprocta semicincta. The scale bar represents 10 µm. authors also noticed that Candidatus Image reproduced from McCutcheon, J. P., McDonald, B. R. & Moran, N. P. Origin of an alternative genetic Hodgkinia cicadicola uses an code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 5, e1000565 (2009). alternative genetic code. In most species the codon UGA is a stop Such recoding events are rare and genome, McCutcheon et al.