Lamellar Bodies of Human Epidermis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Transcriptome Response of High- and Low-Light-Adapted Prochlorococcus Strains to Changing Iron Availability
Transcriptome response of high- and low-light-adapted Prochlorococcus strains to changing iron availability The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation Thompson, Anne W et al. “Transcriptome Response of High- and Low-light-adapted Prochlorococcus Strains to Changing Iron Availability.” ISME Journal (2011), 1-15. As Published http://dx.doi.org/10.1038/ismej.2011.49 Publisher Nature Publishing Group Version Author's final manuscript Citable link http://hdl.handle.net/1721.1/64705 Terms of Use Creative Commons Attribution-Noncommercial-Share Alike 3.0 Detailed Terms http://creativecommons.org/licenses/by-nc-sa/3.0/ Title: Transcriptome response of high- and low-light adapted Prochlorococcus strains to changing iron availability Running title: Prochlorococcus response to iron stress 5 Contributors: Anne W. Thompson1, Katherine Huang1, Mak A. Saito* 2, Sallie W. Chisholm* 1, 3 10 1 MIT Department of Civil and Environmental Engineering 2 Woods Hole Oceanographic Institution – Department of Marine Chemistry and Geochemistry 3 MIT Department of Biology 15 * To whom correspondence should be addressed: E-mail: [email protected] and [email protected] Subject Category: Microbial population and community ecology 20 Abstract Prochlorococcus contributes significantly to ocean primary productivity. The link between primary productivity and iron in specific ocean regions is well established and iron-limitation of Prochlorococcus cell division rates in these regions has been 25 demonstrated. However, the extent of ecotypic variation in iron metabolism among Prochlorococcus and the molecular basis for differences is not understood. Here, we examine the growth and transcriptional response of Prochlorococcus strains, MED4 and MIT9313, to changing iron concentrations. -

Germline Mutations Affecting Gα11 in Hypoparathyroidism
T h e new england journal o f medicine of a meta-analysis of studies of the effect of re- terone levels were increased. Although long- duced dietary salt on the incidence of cardiovas- term, modest reductions in salt intake result in cular events and death.1 The authors of the Co- small, physiologic increases in plasma renin ac- chrane report wrote that there was “no strong tivity,3 the preponderance of evidence suggests evidence of benefit.” In a summary statement, we that a reduced salt intake is associated with a wrote that this particular Cochrane analysis con- decreased risk of cardiovascular events and cluded that reducing dietary salt intake did not death. Furthermore, it is worth remembering decrease the risk of death or cardiovascular dis- that diuretics remain one of the most effective ease. Stigler et al. suggest that “indeterminate antihypertensive therapies, and their beneficial results,” rather than no significant effect, would effect on cardiovascular disease is well docu- be a more appropriate interpretation of the analy- mented.4 Nevertheless, as we suggested, in sis. Both interpretations may be correct. Although terms of safety, the lower limit of salt consump- it may not be possible to reject the null hypothe- tion has not been clearly defined. sis with certainty (i.e., no effect of reduced salt), Theodore A. Kotchen, M.D. the analysis should have been powered to detect Allen W. Cowley, Jr., Ph.D. a clinically meaningful difference, and the con- Medical College of Wisconsin clusion of no effect provides more guidance than Milwaukee, WI “indeterminate results” to clinical decision mak- [email protected] ers. -

Differential Expression Profiling of Gene Response to Ionizing Radiation in Two Endometrial Cancer Cell Lines with Distinct Radiosensitivities
625-634 28/1/2009 12:32 ÌÌ ™ÂÏ›‰·625 ONCOLOGY REPORTS 21: 625-634, 2009 625 Differential expression profiling of gene response to ionizing radiation in two endometrial cancer cell lines with distinct radiosensitivities XUE-LIAN DU1,2, TAO JIANG2, ZE-QING WEN1, QING-SHUI LI2, RONG GAO2 and FEI WANG1 1Department of Gynecologic Oncology, Shandong Tumor Hospital, Shandong University, Jinan 250117; 2Department of Obstetrics and Gynecology, Provincial Hospital Affiliated to Shandong University, Jinan 250021, P.R. China Received November 4, 2008; Accepted December 17, 2008 DOI: 10.3892/or_00000265 Abstracts. Although radiotherapy is routinely administered Introduction to high-risk endometrial carcinoma and offer a significant disease-free survival advantage, the therapeutic effect is Endometrial cancer is one of the most common gynecological sometimes limited by the occurrence of radioresistance. To malignancies worldwide. Surgery is the preferred initial determine the patterns of gene expression responsible for treatment and most women with early-stage, low-risk disease the radioresistance and to search for potential target genes for will do well without adjuvant radiotherapy. However, both radiotherapy, we selected two cell lines with distinct radio- intermediate-risk and high-risk patients are at risk for local- sensitivities using colony-formation assay from four endo- regional relapse and therefore adjuvant radiotherapy, such as metrial cancer cell lines. The cell cycle distribution showed pelvic radiation, vaginal brachytherapy, and whole-abdomen higher fractions of G2/M phase cells in the radiosensitive cell radiation, is essential for local control (1). Johnson and line KLE after radiation compared with the radioresistant cell colleagues reported that adjuvant external-beam pelvic radio- line ISK. -

Sphingolipid Metabolism Diseases ⁎ Thomas Kolter, Konrad Sandhoff
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Biochimica et Biophysica Acta 1758 (2006) 2057–2079 www.elsevier.com/locate/bbamem Review Sphingolipid metabolism diseases ⁎ Thomas Kolter, Konrad Sandhoff Kekulé-Institut für Organische Chemie und Biochemie der Universität, Gerhard-Domagk-Str. 1, D-53121 Bonn, Germany Received 23 December 2005; received in revised form 26 April 2006; accepted 23 May 2006 Available online 14 June 2006 Abstract Human diseases caused by alterations in the metabolism of sphingolipids or glycosphingolipids are mainly disorders of the degradation of these compounds. The sphingolipidoses are a group of monogenic inherited diseases caused by defects in the system of lysosomal sphingolipid degradation, with subsequent accumulation of non-degradable storage material in one or more organs. Most sphingolipidoses are associated with high mortality. Both, the ratio of substrate influx into the lysosomes and the reduced degradative capacity can be addressed by therapeutic approaches. In addition to symptomatic treatments, the current strategies for restoration of the reduced substrate degradation within the lysosome are enzyme replacement therapy (ERT), cell-mediated therapy (CMT) including bone marrow transplantation (BMT) and cell-mediated “cross correction”, gene therapy, and enzyme-enhancement therapy with chemical chaperones. The reduction of substrate influx into the lysosomes can be achieved by substrate reduction therapy. Patients suffering from the attenuated form (type 1) of Gaucher disease and from Fabry disease have been successfully treated with ERT. © 2006 Elsevier B.V. All rights reserved. Keywords: Ceramide; Lysosomal storage disease; Saposin; Sphingolipidose Contents 1. Sphingolipid structure, function and biosynthesis ..........................................2058 1.1. -

Transcriptomic Analysis of the Aquaporin (AQP) Gene Family
Pancreatology 19 (2019) 436e442 Contents lists available at ScienceDirect Pancreatology journal homepage: www.elsevier.com/locate/pan Transcriptomic analysis of the Aquaporin (AQP) gene family interactome identifies a molecular panel of four prognostic markers in patients with pancreatic ductal adenocarcinoma Dimitrios E. Magouliotis a, b, Vasiliki S. Tasiopoulou c, Konstantinos Dimas d, * Nikos Sakellaridis d, Konstantina A. Svokos e, Alexis A. Svokos f, Dimitris Zacharoulis b, a Division of Surgery and Interventional Science, Faculty of Medical Sciences, UCL, London, UK b Department of Surgery, University of Thessaly, Biopolis, Larissa, Greece c Faculty of Medicine, School of Health Sciences, University of Thessaly, Biopolis, Larissa, Greece d Department of Pharmacology, Faculty of Medicine, School of Health Sciences, University of Thessaly, Biopolis, Larissa, Greece e The Warren Alpert Medical School of Brown University, Providence, RI, USA f Riverside Regional Medical Center, Newport News, VA, USA article info abstract Article history: Background: This study aimed to assess the differential gene expression of aquaporin (AQP) gene family Received 14 October 2018 interactome in pancreatic ductal adenocarcinoma (PDAC) using data mining techniques to identify novel Received in revised form candidate genes intervening in the pathogenicity of PDAC. 29 January 2019 Method: Transcriptome data mining techniques were used in order to construct the interactome of the Accepted 9 February 2019 AQP gene family and to determine which genes members are differentially expressed in PDAC as Available online 11 February 2019 compared to controls. The same techniques were used in order to evaluate the potential prognostic role of the differentially expressed genes. Keywords: PDAC Results: Transcriptome microarray data of four GEO datasets were incorporated, including 142 primary Aquaporin tumor samples and 104 normal pancreatic tissue samples. -

A Minimum-Labeling Approach for Reconstructing Protein Networks Across Multiple Conditions
A Minimum-Labeling Approach for Reconstructing Protein Networks across Multiple Conditions Arnon Mazza1, Irit Gat-Viks2, Hesso Farhan3, and Roded Sharan1 1 Blavatnik School of Computer Science, Tel Aviv University, Tel Aviv 69978, Israel. Email: [email protected]. 2 Dept. of Cell Research and Immunology, Tel Aviv University, Tel Aviv 69978, Israel. 3 Biotechnology Institute Thurgau, University of Konstanz, Unterseestrasse 47, CH-8280 Kreuzlingen, Switzerland. Abstract. The sheer amounts of biological data that are generated in recent years have driven the development of network analysis tools to fa- cilitate the interpretation and representation of these data. A fundamen- tal challenge in this domain is the reconstruction of a protein-protein sub- network that underlies a process of interest from a genome-wide screen of associated genes. Despite intense work in this area, current algorith- mic approaches are largely limited to analyzing a single screen and are, thus, unable to account for information on condition-specific genes, or reveal the dynamics (over time or condition) of the process in question. Here we propose a novel formulation for network reconstruction from multiple-condition data and devise an efficient integer program solution for it. We apply our algorithm to analyze the response to influenza in- fection in humans over time as well as to analyze a pair of ER export related screens in humans. By comparing to an extant, single-condition tool we demonstrate the power of our new approach in integrating data from multiple conditions in a compact and coherent manner, capturing the dynamics of the underlying processes. 1 Introduction With the increasing availability of high-throughput data, network biol- arXiv:1307.7803v1 [q-bio.QM] 30 Jul 2013 ogy has become the method of choice for filtering, interpreting and rep- resenting these data. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Structural Evidence for Adaptive Ligand Binding of Glycolipid Transfer Protein
doi:10.1016/j.jmb.2005.10.031 J. Mol. Biol. (2006) 355, 224–236 Structural Evidence for Adaptive Ligand Binding of Glycolipid Transfer Protein Tomi T. Airenne†, Heidi Kidron†, Yvonne Nymalm, Matts Nylund Gun West, Peter Mattjus and Tiina A. Salminen* Department of Biochemistry Glycolipids participate in many important cellular processes and they are and Pharmacy, A˚ bo Akademi bound and transferred with high specificity by glycolipid transfer protein University, Tykisto¨katu 6A (GLTP). We have solved three different X-ray structures of bovine GLTP at FIN-20520 Turku, Finland 1.4 A˚ , 1.6 A˚ and 1.8 A˚ resolution, all with a bound fatty acid or glycolipid. The 1.4 A˚ structure resembles the recently characterized apo-form of the human GLTP but the other two structures represent an intermediate conformation of the apo-GLTPs and the human lactosylceramide-bound GLTP structure. These novel structures give insight into the mechanism of lipid binding and how GLTP may conformationally adapt to different lipids. Furthermore, based on the structural comparison of the GLTP structures and the three-dimensional models of the related Podospora anserina HET-C2 and Arabidopsis thaliana accelerated cell death protein, ACD11, we give structural explanations for their specific lipid binding properties. q 2005 Elsevier Ltd. All rights reserved. Keywords: crystal structure; homology modeling; conformational change; *Corresponding author cavity; fluorescence Introduction to the diverse roles of glycolipids in the cell, GLTP could potentially function as a modulator -

Glucocorticoids Accelerate Fetal Maturation of the Epidermal Permeability Barrier in the Rat
Glucocorticoids accelerate fetal maturation of the epidermal permeability barrier in the rat. M Aszterbaum, … , G K Menon, M L Williams J Clin Invest. 1993;91(6):2703-2708. https://doi.org/10.1172/JCI116509. Research Article The cutaneous permeability barrier to systemic water loss is mediated by hydrophobic lipids forming membrane bilayers within the intercellular domains of the stratum corneum (SC). The barrier emerges during day 20 of gestation in the fetal rat and is correlated with increasing SC thickness and increasing SC lipid content, the appearance of well-formed lamellar bodies in the epidermis, and the presence of lamellar unit structures throughout the SC. Because glucocorticoids accelerate lung lamellar body and surfactant maturation in man and experimental animals, these studies were undertaken to determine whether maternal glucocorticoid treatment accelerates maturation of the epidermal lamellar body secretory system. Maternal rats were injected with betamethasone or saline (control) on days 16-18, and pups were delivered prematurely on day 19. Whereas control pups exhibited immature barriers to transepidermal water loss (8.16 +/- 0.52 mg/cm2 per h), glucocorticoid-treated pups exhibited competent barriers (0.74 +/- 0.14 mg/cm2 per h; P < 0.001). Glucocorticoid treatment also: (a) accelerated maturation of lamellar body and SC membrane ultrastructure; (b) increased SC total lipid content twofold; and (c) increased cholesterol and polar ceramide content three- to sixfold. Thus, glucocorticoids accelerate the functional, morphological, and lipid biochemical maturation of the permeability barrier in the fetal rat. Find the latest version: https://jci.me/116509/pdf Glucocorticoids Accelerate Fetal Maturation of the Epidermal Permeability Barrier in the Rat Michelle Aszterbaum, * Kenneth R. -

Ubiquitylome Profiling of Parkin-Null Brain Reveals Dysregulation Of
Neurobiology of Disease 127 (2019) 114–130 Contents lists available at ScienceDirect Neurobiology of Disease journal homepage: www.elsevier.com/locate/ynbdi Ubiquitylome profiling of Parkin-null brain reveals dysregulation of calcium T homeostasis factors ATP1A2, Hippocalcin and GNA11, reflected by altered firing of noradrenergic neurons Key J.a,1, Mueller A.K.b,1, Gispert S.a, Matschke L.b, Wittig I.c, Corti O.d,e,f,g, Münch C.h, ⁎ ⁎ Decher N.b, , Auburger G.a, a Exp. Neurology, Goethe University Medical School, 60590 Frankfurt am Main, Germany b Institute for Physiology and Pathophysiology, Vegetative Physiology and Marburg Center for Mind, Brain and Behavior - MCMBB; Clinic for Neurology, Philipps-University Marburg, 35037 Marburg, Germany c Functional Proteomics, SFB 815 Core Unit, Goethe University Medical School, 60590 Frankfurt am Main, Germany d Institut du Cerveau et de la Moelle épinière, ICM, Paris, F-75013, France e Inserm, U1127, Paris, F-75013, France f CNRS, UMR 7225, Paris, F-75013, France g Sorbonne Universités, Paris, F-75013, France h Institute of Biochemistry II, Goethe University Medical School, 60590 Frankfurt am Main, Germany ARTICLE INFO ABSTRACT Keywords: Parkinson's disease (PD) is the second most frequent neurodegenerative disorder in the old population. Among Parkinson's disease its monogenic variants, a frequent cause is a mutation in the Parkin gene (Prkn). Deficient function of Parkin Mitochondria triggers ubiquitous mitochondrial dysfunction and inflammation in the brain, but it remains unclear howse- Parkin lective neural circuits become vulnerable and finally undergo atrophy. Ubiquitin We attempted to go beyond previous work, mostly done in peripheral tumor cells, which identified protein Calcium targets of Parkin activity, an ubiquitin E3 ligase. -

Conserved and Novel Properties of Clathrin-Mediated Endocytosis in Dictyostelium Discoideum" (2012)
Rockefeller University Digital Commons @ RU Student Theses and Dissertations 2012 Conserved and Novel Properties of Clathrin- Mediated Endocytosis in Dictyostelium Discoideum Laura Macro Follow this and additional works at: http://digitalcommons.rockefeller.edu/ student_theses_and_dissertations Part of the Life Sciences Commons Recommended Citation Macro, Laura, "Conserved and Novel Properties of Clathrin-Mediated Endocytosis in Dictyostelium Discoideum" (2012). Student Theses and Dissertations. Paper 163. This Thesis is brought to you for free and open access by Digital Commons @ RU. It has been accepted for inclusion in Student Theses and Dissertations by an authorized administrator of Digital Commons @ RU. For more information, please contact [email protected]. CONSERVED AND NOVEL PROPERTIES OF CLATHRIN- MEDIATED ENDOCYTOSIS IN DICTYOSTELIUM DISCOIDEUM A Thesis Presented to the Faculty of The Rockefeller University in Partial Fulfillment of the Requirements for the degree of Doctor of Philosophy by Laura Macro June 2012 © Copyright by Laura Macro 2012 CONSERVED AND NOVEL PROPERTIES OF CLATHRIN- MEDIATED ENDOCYTOSIS IN DICTYOSTELIUM DISCOIDEUM Laura Macro, Ph.D. The Rockefeller University 2012 The protein clathrin mediates one of the major pathways of endocytosis from the extracellular milieu and plasma membrane. Clathrin functions with a network of interacting accessory proteins, one of which is the adaptor complex AP-2, to co-ordinate vesicle formation. Disruption of genes involved in clathrin-mediated endocytosis causes embryonic lethality in multicellular animals suggesting that clathrin-mediated endocytosis is a fundamental cellular process. However, loss of clathrin-mediated endocytosis genes in single cell eukaryotes, such as S.cerevisiae (yeast), does not cause lethality, suggesting that clathrin may convey specific advantages for multicellularity. -
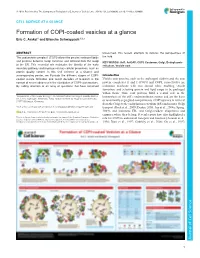
Formation of COPI-Coated Vesicles at a Glance Eric C
© 2018. Published by The Company of Biologists Ltd | Journal of Cell Science (2018) 131, jcs209890. doi:10.1242/jcs.209890 CELL SCIENCE AT A GLANCE Formation of COPI-coated vesicles at a glance Eric C. Arakel1 and Blanche Schwappach1,2,* ABSTRACT unresolved, this review attempts to refocus the perspectives of The coat protein complex I (COPI) allows the precise sorting of lipids the field. and proteins between Golgi cisternae and retrieval from the Golgi KEY WORDS: Arf1, ArfGAP, COPI, Coatomer, Golgi, Endoplasmic to the ER. This essential role maintains the identity of the early reticulum, Vesicle coat secretory pathway and impinges on key cellular processes, such as protein quality control. In this Cell Science at a Glance and accompanying poster, we illustrate the different stages of COPI- Introduction coated vesicle formation and revisit decades of research in the Vesicle coat proteins, such as the archetypal clathrin and the coat context of recent advances in the elucidation of COPI coat structure. protein complexes II and I (COPII and COPI, respectively) are By calling attention to an array of questions that have remained molecular machines with two central roles: enabling vesicle formation, and selecting protein and lipid cargo to be packaged within them. Thus, coat proteins fulfil a central role in the 1Department of Molecular Biology, Universitätsmedizin Göttingen, Humboldtallee homeostasis of the cell’s endomembrane system and are the basis 23, 37073 Göttingen, Germany. 2Max-Planck Institute for Biophysical Chemistry, 37077 Göttingen, Germany. of functionally segregated compartments. COPI operates in retrieval from the Golgi to the endoplasmic reticulum (ER) and in intra-Golgi *Author for correspondence ([email protected]) transport (Beck et al., 2009; Duden, 2003; Lee et al., 2004a; Spang, E.C.A., 0000-0001-7716-7149; B.S., 0000-0003-0225-6432 2009), and maintains ER- and Golgi-resident chaperones and enzymes where they belong.