Application of a Triggered MRM Database and Library for the Quantitation and Identification of Pesticides in Food Extracts
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Environmental Properties of Chemicals Volume 2
1 t ENVIRONMENTAL 1 PROTECTION Esa Nikunen . Riitta Leinonen Birgit Kemiläinen • Arto Kultamaa Environmental properties of chemicals Volume 2 1 O O O O O O O O OO O OOOOOO Ol OIOOO FINNISH ENVIRONMENT INSTITUTE • EDITA Esa Nikunen e Riitta Leinonen Birgit Kemiläinen • Arto Kultamaa Environmental properties of chemicals Volume 2 HELSINKI 1000 OlO 00000001 00000000000000000 Th/s is a second revfsed version of Environmental Properties of Chemica/s, published by VAPK-Pub/ishing and Ministry of Environment, Environmental Protection Department as Research Report 91, 1990. The pubiication is also available as a CD ROM version: EnviChem 2.0, a PC database runniny under Windows operating systems. ISBN 951-7-2967-2 (publisher) ISBN 952-7 1-0670-0 (co-publisher) ISSN 1238-8602 Layout: Pikseri Julkaisupalvelut Cover illustration: Jussi Hirvi Edita Ltd. Helsinki 2000 Environmental properties of chemicals Volume 2 _____ _____________________________________________________ Contents . VOLUME ONE 1 Contents of the report 2 Environmental properties of chemicals 3 Abbreviations and explanations 7 3.1 Ways of exposure 7 3.2 Exposed species 7 3.3 Fffects________________________________ 7 3.4 Length of exposure 7 3.5 Odour thresholds 8 3.6 Toxicity endpoints 9 3.7 Other abbreviations 9 4 Listofexposedspecies 10 4.1 Mammais 10 4.2 Plants 13 4.3 Birds 14 4.4 Insects 17 4.5 Fishes 1$ 4.6 Mollusca 22 4.7 Crustaceans 23 4.8 Algae 24 4.9 Others 25 5 References 27 Index 1 List of chemicals in alphabetical order - 169 Index II List of chemicals in CAS-number order -

Guide No. 1 – October 2020 2/12 the CONCEPT and IMPLEMENTATION of CPA GUIDANCE RESIDUE LEVELS
Cooperation Centre for Scientific Research Relative to Tobacco CORESTA GUIDE N° 1 The Concept and Implementation of CPA Guidance Residue Levels October 2020 Agro-Chemical Advisory Committee CORESTA TECHNICAL GUIDE N° 1 Title: The Concept and Implementation of CPA Guidance Residue Levels Status: Valid Note: This document will be periodically reviewed by CORESTA Document history: Date of review Information July 2003 Version 1 GRL for Pyrethrins () and Terbufos corrected. December 2003 CPA terminology corrected. June 2008 Version 2 – GRLs revised and residue definitions added Provisional GRL of 2.00 ppm for Cyfluthrin to replace previous June 2010 GRL of 0.50 ppm July 2013 Version 3 – GRLs revised October 2013 Note for Maleic Hydrazide revised Version 4 – GRLs revised + clarification that scope of GRLs July 2016 applies predominantly to the production of traditional cigarette tobaccos and GAP associated with their cultivation. June 2018 Fluopyram GRL of 5 ppm added to GRL list Version 5 – Nine new CPAs with GRL added to list. November 2019 Revision of GRLs for Chlorantraniliprole and Indoxacarb. Updated web links. October 2020 Version 6 – Flupyradifurone GRL of 21 ppm added to GRL list. CORESTA Guide No. 1 – October 2020 2/12 THE CONCEPT AND IMPLEMENTATION OF CPA GUIDANCE RESIDUE LEVELS Executive Summary • Guidance Residue Levels (GRLs) are in the remit of the Agro-Chemical Advisory Committee (ACAC) of CORESTA. Their development is a joint activity of all ACAC members, who represent the leaf production, processing and manufacturing sectors of the Tobacco Industry. The concept of GRLs and their implementation are described in this guide. • GRLs provide guidance to tobacco growers and assist with interpretation and evaluation of results from analyses of residues of Crop Protection Agents (CPAs*). -
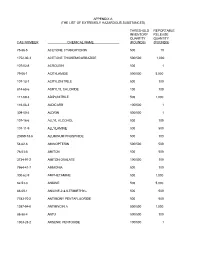
The List of Extremely Hazardous Substances)
APPENDIX A (THE LIST OF EXTREMELY HAZARDOUS SUBSTANCES) THRESHOLD REPORTABLE INVENTORY RELEASE QUANTITY QUANTITY CAS NUMBER CHEMICAL NAME (POUNDS) (POUNDS) 75-86-5 ACETONE CYANOHYDRIN 500 10 1752-30-3 ACETONE THIOSEMICARBAZIDE 500/500 1,000 107-02-8 ACROLEIN 500 1 79-06-1 ACRYLAMIDE 500/500 5,000 107-13-1 ACRYLONITRILE 500 100 814-68-6 ACRYLYL CHLORIDE 100 100 111-69-3 ADIPONITRILE 500 1,000 116-06-3 ALDICARB 100/500 1 309-00-2 ALDRIN 500/500 1 107-18-6 ALLYL ALCOHOL 500 100 107-11-9 ALLYLAMINE 500 500 20859-73-8 ALUMINUM PHOSPHIDE 500 100 54-62-6 AMINOPTERIN 500/500 500 78-53-5 AMITON 500 500 3734-97-2 AMITON OXALATE 100/500 100 7664-41-7 AMMONIA 500 100 300-62-9 AMPHETAMINE 500 1,000 62-53-3 ANILINE 500 5,000 88-05-1 ANILINE,2,4,6-TRIMETHYL- 500 500 7783-70-2 ANTIMONY PENTAFLUORIDE 500 500 1397-94-0 ANTIMYCIN A 500/500 1,000 86-88-4 ANTU 500/500 100 1303-28-2 ARSENIC PENTOXIDE 100/500 1 THRESHOLD REPORTABLE INVENTORY RELEASE QUANTITY QUANTITY CAS NUMBER CHEMICAL NAME (POUNDS) (POUNDS) 1327-53-3 ARSENOUS OXIDE 100/500 1 7784-34-1 ARSENOUS TRICHLORIDE 500 1 7784-42-1 ARSINE 100 100 2642-71-9 AZINPHOS-ETHYL 100/500 100 86-50-0 AZINPHOS-METHYL 10/500 1 98-87-3 BENZAL CHLORIDE 500 5,000 98-16-8 BENZENAMINE, 3-(TRIFLUOROMETHYL)- 500 500 100-14-1 BENZENE, 1-(CHLOROMETHYL)-4-NITRO- 500/500 500 98-05-5 BENZENEARSONIC ACID 10/500 10 3615-21-2 BENZIMIDAZOLE, 4,5-DICHLORO-2-(TRI- 500/500 500 FLUOROMETHYL)- 98-07-7 BENZOTRICHLORIDE 100 10 100-44-7 BENZYL CHLORIDE 500 100 140-29-4 BENZYL CYANIDE 500 500 15271-41-7 BICYCLO[2.2.1]HEPTANE-2-CARBONITRILE,5- -

Application of Dried Blood Spots for the Determination of Organophosphorus Pesticides by GC-MS/MS
UNIVERSIDADE DA BEIRA INTERIOR Ciências Application of dried blood spots for the determination of organophosphorus pesticides by GC-MS/MS Sofia Pires Seixo Soares Dissertação para obtenção do Grau de Mestre em Bioquímica (2º ciclo de estudos) Orientador: Doutor Mário Jorge Dinis Barroso Co-orientador: Professora Doutora Maria Eugenia Gallardo Alba Covilhã, junho de 2018 Agradecimentos A realização de uma dissertação de mestrado é o culminar de muitas horas de estudo, reflexão e trabalho pessoal, mas representa também uma etapa que não seria possível sem o contributo fundamental de várias pessoas. À Doutora Eugénia Gallardo e ao Doutor Mário Barroso, como excelentes mentores que são, quero agradecer toda a orientação, motivação, incentivo e confiança. Toda a ajuda e partilha de experiência e conhecimento durante todo este percurso. Ao Tiago Rosado, pela imprescindível ajuda e disponibilidade constante, bem como pela amizade. À Joana Gonçalves e ao Ângelo Luís um agradecimento especial pela amizade e carinho demonstrado ao longo deste ano. A todos, quero agradecer pelo bom ambiente proporcionado. Aos meus pais Ana Soares e José Soares, a quem dedico este feito, por me darem a possibilidade de concretizar conquistas como esta, pelo apoio incondicional, por sempre terem acreditado em mim e por fazerem parte deste momento. Aos meus avós e restante família, por todo o apoio e preocupação. Um agradecimento particular à Inês Ramos, à Rita Marques, à Maria Cunha e ao Pedro Batista pelo companheirismo, amizade e apoio de há tantos anos. E por último, mas nunca menos importante, um enorme obrigada ao meu namorado António Fernandes por toda a paciência, pelo incansável apoio, carinho e ajuda. -

Organophosphate Insecticides
CHAPTER 4 HIGHLIGHTS Organophosphate Insecticides Acts through phosphorylation of the acetylcholinesterase enzyme Since the removal of organochlorine insecticides from use, organophosphate at nerve endings insecticides have become the most widely used insecticides available today. More Absorbed by inhalation, than forty of them are currently registered for use and all run the risk of acute ingestion, and skin and subacute toxicity. Organophosphates are used in agriculture, in the home, penetration in gardens, and in veterinary practice. All apparently share a common mecha- Muscarinic, nicotinic & CNS nism of cholinesterase inhibition and can cause similar symptoms. Because they effects share this mechanism, exposure to the same organophosphate by multiple routes or to multiple organophosphates by multiple routes can lead to serious additive Signs and Symptoms: toxicity. It is important to understand, however, that there is a wide range of Headache, hypersecretion, toxicity in these agents and wide variation in cutaneous absorption, making muscle twitching, nausea, specific identification and management quite important. diarrhea Respiratory depression, seizures, loss of consciousness Toxicology Miosis is often a helpful Organophosphates poison insects and mammals primarily by phosphory- diagnostic sign lation of the acetylcholinesterase enzyme (AChE) at nerve endings. The result is a loss of available AChE so that the effector organ becomes overstimulated by Treatment: the excess acetylcholine (ACh, the impulse-transmitting substance) in the nerve Clear airway, improve tissue ending. The enzyme is critical to normal control of nerve impulse transmission oxygenation from nerve fibers to smooth and skeletal muscle cells, glandular cells, and Administer atropine sulfate autonomic ganglia, as well as within the central nervous system (CNS). -

Investigation on the Behavior of Pesticides in Atmosphere
Aerosol and Air Quality Research, 11: 783–790, 2011 Copyright © Taiwan Association for Aerosol Research ISSN: 1680-8584 print / 2071-1409 online doi: 10.4209/aaqr.2010.10.0085 Investigation on the Behavior of Pesticides in Atmosphere Pasquale Avino1, Giuseppe Cinelli2, Ivan Notardonato2, Mario V. Russo2* 1 DIPIA, INAIL (ex-ISPESL), via Urbana 167, 00184 Rome, Italy 2 Faculty of Agriculture, University of Molise, via De Sanctis, Campobasso, Italy ABSTRACT Although pesticides are widely used in agriculture, they and in particular the relative residues in foodstuffs, water and atmosphere, may cause remarkable sanitary problems due to the harmful effects (carcinogenic and mutagenic effects) on the human health. In fact, their spread in waters and atmosphere can produce undesired effects on various organisms and/or water contamination. This paper shows an analytical approach based on XAD-2 adsorbent and GC analysis for evaluating the pesticide trend in atmosphere: in particular, the pesticides investigated are omethoate, dicrotofos, disulfoton, dimethoate, parathion methyl, formothion, paraoxon ethyl, malaoxon, parathion ethyl, iodofenfos and triazofos. For the analytical methodology a linearity response was obtained (r2 = 0.9988) in GC-NPD whereas the limits of detection range between 2 and 5 pg/μL in GC-NPD with a Relative Standard Deviation below 9.5. Finally, this approach has been successfully applied to real samples: the results show that dimethoate concentration decreases with increasing distance from the sampling site but it is still persistent in atmosphere after few days from the pesticide spraying. Keywords: Pesticides; XAD-2 adsorbent; GC analysis; Atmosphere; Air quality. INTRODUCTION 2009). One of the most important OP reactions is water hydrolysis. -

Recommended Classification of Pesticides by Hazard and Guidelines to Classification 2019 Theinternational Programme on Chemical Safety (IPCS) Was Established in 1980
The WHO Recommended Classi cation of Pesticides by Hazard and Guidelines to Classi cation 2019 cation Hazard of Pesticides by and Guidelines to Classi The WHO Recommended Classi The WHO Recommended Classi cation of Pesticides by Hazard and Guidelines to Classi cation 2019 The WHO Recommended Classification of Pesticides by Hazard and Guidelines to Classification 2019 TheInternational Programme on Chemical Safety (IPCS) was established in 1980. The overall objectives of the IPCS are to establish the scientific basis for assessment of the risk to human health and the environment from exposure to chemicals, through international peer review processes, as a prerequisite for the promotion of chemical safety, and to provide technical assistance in strengthening national capacities for the sound management of chemicals. This publication was developed in the IOMC context. The contents do not necessarily reflect the views or stated policies of individual IOMC Participating Organizations. The Inter-Organization Programme for the Sound Management of Chemicals (IOMC) was established in 1995 following recommendations made by the 1992 UN Conference on Environment and Development to strengthen cooperation and increase international coordination in the field of chemical safety. The Participating Organizations are: FAO, ILO, UNDP, UNEP, UNIDO, UNITAR, WHO, World Bank and OECD. The purpose of the IOMC is to promote coordination of the policies and activities pursued by the Participating Organizations, jointly or separately, to achieve the sound management of chemicals in relation to human health and the environment. WHO recommended classification of pesticides by hazard and guidelines to classification, 2019 edition ISBN 978-92-4-000566-2 (electronic version) ISBN 978-92-4-000567-9 (print version) ISSN 1684-1042 © World Health Organization 2020 Some rights reserved. -

United States Patent (19) 11 Patent Number: 5,703,064 Yokoi Et Al
US005703064A United States Patent (19) 11 Patent Number: 5,703,064 Yokoi et al. 45) Date of Patent: Dec. 30, 1997 54 PESTICIDAL COMBINATIONS FOREIGN PATENT DOCUMENTS 75 Inventors: Shinji Yokoi; Akira Nishida, both of 0 196524 10/1986 European Pat. Of.. Shiga-ken; Tokio Obata; Kouichi Golka, both of Ube, all of Japan OTHER PUBLICATIONS 73) Assignees: Sankyo Company, Limited, Tokyo; Worthing et al, The Pesticide Manual, 9th Ed. (1991), pp. Ube industries Ltd., Ube, both of 747 and 748. Japan L.C. Gaughan et al., "Pesticide interactions: effects of orga nophosphorus pesticides on the metabolism, toxicity, and 21 Appl. No.: 405,795 persistence of selected pyrethroid insecticides". Chemical Abstracts, vol. 94, No. 9, 1981, No. 59740k of Pestic. 22 Filed: Mar 16, 1995 Biochem. Physio., vol. 14, No. 1, 1980, pp. 81-85. 30 Foreign Application Priority Data I. Ishaaya et al., "Cypermethrin synergism by pyrethroid esterase inhibitors in adults of the whitefly Bemisia tabaci". Mar 16, 1994 JP Japan ............................ HE6045.405 Chemical Abstracts, vol. 107, No. 9, 1987, No. 72818y of (51) Int. Cl................. A01N 43/54; A01N 57/00 Pestic Biochem. Physiol., vol. 28, No. 2, 1987, pp. 155-162. 52 U.S. C. ......................................... 51480; 514/256 (58) Field of Search ..................................... 514/80, 256 Primary Examiner Allen J. Robinson Attorney, Agent, or Firm-Frishauf, Holtz, Goodman, 56 References Cited Langer & Chick, P.C. U.S. PATENT DOCUMENTS 57 ABSTRACT 4,374,833 2/1983 Badmin et al. ...................... 424/225 Combinations of the known compound pyrimidifen with 4,845,097 7/1989 Matsumoto et al... 514/234.2 phosphorus-containing pesticides have a synergistic pesti 4,935,516 6/1990 Ataka et al. -

List of Lists
United States Office of Solid Waste EPA 550-B-10-001 Environmental Protection and Emergency Response May 2010 Agency www.epa.gov/emergencies LIST OF LISTS Consolidated List of Chemicals Subject to the Emergency Planning and Community Right- To-Know Act (EPCRA), Comprehensive Environmental Response, Compensation and Liability Act (CERCLA) and Section 112(r) of the Clean Air Act • EPCRA Section 302 Extremely Hazardous Substances • CERCLA Hazardous Substances • EPCRA Section 313 Toxic Chemicals • CAA 112(r) Regulated Chemicals For Accidental Release Prevention Office of Emergency Management This page intentionally left blank. TABLE OF CONTENTS Page Introduction................................................................................................................................................ i List of Lists – Conslidated List of Chemicals (by CAS #) Subject to the Emergency Planning and Community Right-to-Know Act (EPCRA), Comprehensive Environmental Response, Compensation and Liability Act (CERCLA) and Section 112(r) of the Clean Air Act ................................................. 1 Appendix A: Alphabetical Listing of Consolidated List ..................................................................... A-1 Appendix B: Radionuclides Listed Under CERCLA .......................................................................... B-1 Appendix C: RCRA Waste Streams and Unlisted Hazardous Wastes................................................ C-1 This page intentionally left blank. LIST OF LISTS Consolidated List of Chemicals -

NMP-Free Formulations of Neonicotinoids
(19) & (11) EP 2 266 400 A1 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 29.12.2010 Bulletin 2010/52 A01N 43/40 (2006.01) A01N 43/86 (2006.01) A01N 47/40 (2006.01) A01N 51/00 (2006.01) (2006.01) (2006.01) (21) Application number: 09305544.0 A01P 7/00 A01N 25/02 (22) Date of filing: 15.06.2009 (84) Designated Contracting States: (72) Inventors: AT BE BG CH CY CZ DE DK EE ES FI FR GB GR • Gasse, Jean-Jacques HR HU IE IS IT LI LT LU LV MC MK MT NL NO PL 27600 Saint-Aubin-Sur-Gaillon (FR) PT RO SE SI SK TR • Duchamp, Guillaume Designated Extension States: 92230 Gennevilliers (FR) AL BA RS • Cantero, Maria 92230 Gennevilliers (FR) (71) Applicant: NUFARM 92233 Gennevelliers (FR) (74) Representative: Cabinet Plasseraud 52, rue de la Victoire 75440 Paris Cedex 09 (FR) (54) NMP-free formulations of neonicotinoids (57) The invention relates to NMP-free liquid formulation comprising at least one nicotinoid and at least one aprotic polar component selected from the group comprising the compounds of formula I, II or III below, and mixtures thereof, wherein R1 and R2 independently represent H or an alkyl group having less than 5 carbons, preferably a methyl group, and n represents an integer ranging from 0 to 5, and to their applications. EP 2 266 400 A1 Printed by Jouve, 75001 PARIS (FR) EP 2 266 400 A1 Description Technical Field of the invention 5 [0001] The invention relates to novel liquid formulations of neonicotinoids and to their use for treating plants, for protecting plants from pests and/or for controlling pests infestation. -

Ehab Shaaban Mohamed EL-Osely M.B., B
INFLUENCE OF SOME PESTICIDES ON HUMORAL AND CELLULAR IMMUNITY OF EXPOSED WORKERS IN PESTICIDES INDUSTRIES By Ehab Shaaban Mohamed EL-Osely M.B., B. Ch. Faculty of Medicine, Ain Shams university1994 Master in radiodiagnosis, Faculty of Medicine, Ain Shams university1998 A Thesis Submitted in Partial Fulfillment of The Requirement for the Doctor of Philosophy Degree In Environmental Science Department of Medical Science Institute of Environmental Studies & Research Ain Shams University Under The Supervision of: 1- Prof. Dr. Mostafa Mohamed EL Rasad Prof. of Biochemistry Faculty of Medicine, Ain Shams University 2- Dr. Mahmoud Lotfy Sakr Associate Prof. of Forensic Medicine and Toxicology Faculty of Medicine, Ain Shams University 3- Prof. Dr. Zidan Hindy Abdel Hamid Zidan Prof. of Pesticides Chemistry and Toxicology Faculty of Agriculture, Ain Shams University 4- Dr. Amany Mohamed Ezz El-Din Associate Prof. in Radiation Health Research Department National Center for Radiation Research and Technology 5- Dr. Inas Mohamed Fawzy Gaballah Associate Prof. of Industrial medicine Faculty of Medicine, Cairo University 2010 - 1 - Acknowledgement First and foremost thanks to ALLAH to whom I relate any success in achieving any work in my life. I would like to express my deepest appreciation and gratitude to the sole of Prof. Dr. Mostafa Mohamed EL- Rasad, Professor of Biochemistry, Faculty of Medicine Ain Shams University, for suggesting and planning this study. No words can express my deepest thanks to him. My sincere appreciation and special thanks to assistant professor Dr. Mahmoud Lotfy Sakr, assistant professor of clinical toxicology, Faculty of Medicine Ain Shams University, for his overwhelming kindness and encouragement, his keen supervision, continuous guidance, experienced advice. -

Environmental Protection Agency Pt. 355, App. A
Environmental Protection Agency Pt. 355, App. A Release means any spilling, leaking, the facility is located. In the absence pumping, pouring, emitting, emptying, of a SERC for a State or Indian Tribe, discharging, injecting, escaping, leach- the Governor or the chief executive of- ing, dumping, or disposing into the en- ficer of the tribe, respectively, shall be vironment (including the abandonment the SERC. Where there is a cooperative or discarding of barrels, containers, agreement between a State and a and other closed receptacles) of any Tribe, the SERC shall be the entity hazardous chemical, EHS, or CERCLA identified in the agreement. hazardous substance. Solution means any aqueous or or- Reportable quantity means, for any ganic solutions, slurries, viscous solu- CERCLA hazardous substance, the tions, suspensions, emulsions, or quantity established in Table 302.4 of 40 pastes. CFR 302.4, for such substance. For any State means any State of the United EHS, reportable quantity means the States, the District of Columbia, the quantity established in Appendices A Commonwealth of Puerto Rico, Guam, and B of this part for such substance. American Samoa, the United States Unless and until superseded by regula- Virgin Islands, the Northern Mariana tions establishing a reportable quan- Islands, any other territory or posses- tity for newly listed EHSs or CERCLA sion over which the United States has hazardous substances, a weight of 1 jurisdiction and Indian Country. pound shall be the reportable quantity. Threshold planning quantity means, SERC means the State Emergency for a substance listed in Appendices A Response Commission for the State in and B of this part, the quantity listed which the facility is located except in the column ‘‘threshold planning where the facility is located in Indian quantity’’ for that substance.