Tension Suppresses Aurora B Kinase-Triggered Release of Reconstituted Kinetochore-Microtubule Attachments
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

RASSF1A Interacts with and Activates the Mitotic Kinase Aurora-A
Oncogene (2008) 27, 6175–6186 & 2008 Macmillan Publishers Limited All rights reserved 0950-9232/08 $32.00 www.nature.com/onc ORIGINAL ARTICLE RASSF1A interacts with and activates the mitotic kinase Aurora-A L Liu1, C Guo1, R Dammann2, S Tommasi1 and GP Pfeifer1 1Division of Biology, Beckman Research Institute, City of Hope Cancer Center, Duarte, CA, USA and 2Institute of Genetics, University of Giessen, Giessen, Germany The RAS association domain family 1A (RASSF1A) gene tumorigenesis and carcinogen-induced tumorigenesis is located at chromosome 3p21.3 within a specific area of (Tommasi et al., 2005; van der Weyden et al., 2005), common heterozygous and homozygous deletions. RASS- supporting the notion that RASSF1A is a bona fide F1A frequently undergoes promoter methylation-asso- tumor suppressor. However, it is not fully understood ciated inactivation in human cancers. Rassf1aÀ/À mice how RASSF1A is involved in tumor suppression. are prone to both spontaneous and carcinogen-induced The biochemical function of the RASSF1A protein is tumorigenesis, supporting the notion that RASSF1A is a largely unknown. The homology of RASSF1A with the tumor suppressor. However, it is not fully understood how mammalian Ras effector novel Ras effector (NORE)1 RASSF1A is involved in tumor suppression pathways. suggests that the RASSF1A gene product may function Here we show that overexpression of RASSF1A inhibits in signal transduction pathways involving Ras-like centrosome separation. RASSF1A interacts with Aurora-A, proteins. However, recent data indicate that RASSF1A a mitotic kinase. Surprisingly, knockdown of RASS- itself binds to RAS only weakly and that binding to F1A by siRNA led to reduced activation of Aurora-A, RAS may require heterodimerization of RASSF1A and whereas overexpression of RASSF1A resulted in in- NORE1 (Ortiz-Vega et al., 2002). -

HIPK2 and Extrachromosomal Histone H2B Are Separately Recruited by Aurora-B for Cytokinesis
Oncogene https://doi.org/10.1038/s41388-018-0191-6 ARTICLE HIPK2 and extrachromosomal histone H2B are separately recruited by Aurora-B for cytokinesis 1 1 2 3 2 Laura Monteonofrio ● Davide Valente ● Manuela Ferrara ● Serena Camerini ● Roberta Miscione ● 3 1,2 1 Marco Crescenzi ● Cinzia Rinaldo ● Silvia Soddu Received: 14 November 2017 / Revised: 24 January 2018 / Accepted: 5 February 2018 © The Author(s) 2018. This article is published with open access Abstract Cytokinesis, the final phase of cell division, is necessary to form two distinct daughter cells with correct distribution of genomic and cytoplasmic materials. Its failure provokes genetically unstable states, such as tetraploidization and polyploidization, which can contribute to tumorigenesis. Aurora-B kinase controls multiple cytokinetic events, from chromosome condensation to abscission when the midbody is severed. We have previously shown that HIPK2, a kinase involved in DNA damage response and development, localizes at the midbody and contributes to abscission by phosphorylating extrachromosomal histone H2B at Ser14. Of relevance, HIPK2-defective cells do not phosphorylate H2B 1234567890();,: and do not successfully complete cytokinesis leading to accumulation of binucleated cells, chromosomal instability, and increased tumorigenicity. However, how HIPK2 and H2B are recruited to the midbody during cytokinesis is still unknown. Here, we show that regardless of their direct (H2B) and indirect (HIPK2) binding of chromosomal DNA, both H2B and HIPK2 localize at the midbody independently of nucleic acids. Instead, by using mitotic kinase-specific inhibitors in a spatio-temporal regulated manner, we found that Aurora-B kinase activity is required to recruit both HIPK2 and H2B to the midbody. -

The Aurora B Kinase Activity Is Required for the Maintenance of the Differentiated State of Murine Myoblasts
Cell Death and Differentiation (2009) 16, 321–330 & 2009 Macmillan Publishers Limited All rights reserved 1350-9047/09 $32.00 www.nature.com/cdd The Aurora B kinase activity is required for the maintenance of the differentiated state of murine myoblasts G Amabile1,2, AM D’Alise1, M Iovino1, P Jones3, S Santaguida4, A Musacchio4, S Taylor5 and R Cortese*,1 Reversine is a synthetic molecule capable of inducing dedifferentiation of C2C12, a murine myoblast cell line, into multipotent progenitor cells, which can be redirected to differentiate in nonmuscle cell types under appropriate conditions. Reversine is also a potent inhibitor of Aurora B, a protein kinase required for mitotic chromosome segregation, spindle checkpoint function, cytokinesis and histone H3 phosphorylation, raising the possibility that the dedifferentiation capability of reversine is mediated through the inhibition of Aurora B. Indeed, here we show that several other well-characterized Aurora B inhibitors are capable of dedifferentiating C2C12 myoblasts. Significantly, expressing drug-resistant Aurora B mutants, which are insensitive to reversine block the dedifferentiation process, indicating that Aurora B kinase activity is required to maintain the differentiated state. We show that the inhibition of the spindle checkpoint or cytokinesis per se is not sufficient for dedifferentiation. Rather, our data support a model whereby changes in histone H3 phosphorylation result in chromatin remodeling, which in turn restores the multipotent state. Cell Death and Differentiation (2009) 16, 321–330; doi:10.1038/cdd.2008.156; published online 31 October 2008 Lineage-restricted cells can be reprogramed to a state Evidence is emerging that the role of Aurora B is not of pluripotency by several different manipulations, including restricted to mitosis and cell division. -

Discovery of BPR1K871, a Quinazoline Based
www.impactjournals.com/oncotarget/ Oncotarget, 2016, Vol. 7, (No. 52), pp: 86239-86256 Research Paper Discovery of BPR1K871, a quinazoline based, multi-kinase inhibitor for the treatment of AML and solid tumors: Rational design, synthesis, in vitro and in vivo evaluation Yung Chang Hsu1,*, Mohane Selvaraj Coumar2,*, Wen-Chieh Wang1,*, Hui-Yi Shiao1,*, Yi-Yu Ke1, Wen-Hsing Lin1, Ching-Chuan Kuo1, Chun-Wei Chang1, Fu-Ming Kuo1, Pei-Yi Chen1, Sing-Yi Wang1, An-Siou Li1, Chun-Hwa Chen1, Po-Chu Kuo1, Ching- Ping Chen1, Ming-Hsine Wu1, Chen-Lung Huang1, Kuei-Jung Yen1, Yun-I Chang1, John T.-A. Hsu1, Chiung-Tong Chen1, Teng-Kuang Yeh1, Jen-Shin Song1, Chuan Shih1, Hsing-Pang Hsieh1,3 1Institute of Biotechnology and Pharmaceutical Research, National Health Research Institutes, Zhunan, Taiwan, ROC 2Centre for Bioinformatics, School of Life Sciences, Pondicherry University, Kalapet, Puducherry, India 3Department of Chemistry, National Tsing Hua University, Hsinchu, Taiwan, ROC *These authors contributed equally to this work Correspondence to: Hsing-Pang Hsieh, email: [email protected] Keywords: acute myeloid leukemia, aurora kinase, FLT3, quinazoline, multi-kinase inhibitor Received: July 25, 2016 Accepted: November 07, 2016 Published: November 15, 2016 ABSTRACT The design and synthesis of a quinazoline-based, multi-kinase inhibitor for the treatment of acute myeloid leukemia (AML) and other malignancies is reported. Based on the previously reported furanopyrimidine 3, quinazoline core containing lead 4 was synthesized and found to impart dual FLT3/AURKA inhibition (IC50 = 127/5 nM), as well as improved physicochemical properties. A detailed structure-activity relationship study of the lead 4 allowed FLT3 and AURKA inhibition to be finely tuned, resulting in AURKA selective (5 and 7; 100-fold selective over FLT3), FLT3 selective (13; 30-fold selective over AURKA) and dual FLT3/AURKA selective (BPR1K871; IC50 = 19/22 nM) agents. -

Kinase-Targeted Cancer Therapies: Progress, Challenges and Future Directions Khushwant S
Bhullar et al. Molecular Cancer (2018) 17:48 https://doi.org/10.1186/s12943-018-0804-2 REVIEW Open Access Kinase-targeted cancer therapies: progress, challenges and future directions Khushwant S. Bhullar1, Naiara Orrego Lagarón2, Eileen M. McGowan3, Indu Parmar4, Amitabh Jha5, Basil P. Hubbard1 and H. P. Vasantha Rupasinghe6,7* Abstract The human genome encodes 538 protein kinases that transfer a γ-phosphate group from ATP to serine, threonine, or tyrosine residues. Many of these kinases are associated with human cancer initiation and progression. The recent development of small-molecule kinase inhibitors for the treatment of diverse types of cancer has proven successful in clinical therapy. Significantly, protein kinases are the second most targeted group of drug targets, after the G-protein- coupled receptors. Since the development of the first protein kinase inhibitor, in the early 1980s, 37 kinase inhibitors have received FDA approval for treatment of malignancies such as breast and lung cancer. Furthermore, about 150 kinase-targeted drugs are in clinical phase trials, and many kinase-specific inhibitors are in the preclinical stage of drug development. Nevertheless, many factors confound the clinical efficacy of these molecules. Specific tumor genetics, tumor microenvironment, drug resistance, and pharmacogenomics determine how useful a compound will be in the treatment of a given cancer. This review provides an overview of kinase-targeted drug discovery and development in relation to oncology and highlights the challenges and future potential for kinase-targeted cancer therapies. Keywords: Kinases, Kinase inhibition, Small-molecule drugs, Cancer, Oncology Background Recent advances in our understanding of the fundamen- Kinases are enzymes that transfer a phosphate group to a tal molecular mechanisms underlying cancer cell signaling protein while phosphatases remove a phosphate group have elucidated a crucial role for kinases in the carcino- from protein. -

Epigenetic Regulation of Neurogenesis by Citron Kinase Matthew Irg Genti University of Connecticut - Storrs, [email protected]
University of Connecticut OpenCommons@UConn Doctoral Dissertations University of Connecticut Graduate School 12-10-2015 Epigenetic Regulation of Neurogenesis By Citron Kinase Matthew irG genti University of Connecticut - Storrs, [email protected] Follow this and additional works at: https://opencommons.uconn.edu/dissertations Recommended Citation Girgenti, Matthew, "Epigenetic Regulation of Neurogenesis By Citron Kinase" (2015). Doctoral Dissertations. 933. https://opencommons.uconn.edu/dissertations/933 Epigenetic Regulation Of Neurogenesis By Citron Kinase Matthew J. Girgenti, Ph.D. University of Connecticut, 2015 Patterns of neural progenitor division are controlled by a combination of asymmetries in cell division, extracellular signals, and changes in gene expression programs. In a screen for proteins that complex with citron kinase (CitK), a protein essential to cell division in developing brain, I identified the histone methyltransferase- euchromatic histone-lysine N-methyltransferse 2 (G9a). CitK is present in the nucleus and binds to positions in the genome clustered around transcription start sites of genes involved in neuronal development and differentiation. CitK and G9a co-occupy these genomic positions in S/G2-phase of the cell cycle in rat neural progenitors, and CitK functions with G9a to repress gene expression in several developmentally important genes including CDKN1a, H2afz, and Pou3f2/Brn2. The study indicates a novel function for citron kinase in gene repression, and contributes additional evidence to the hypothesis that mechanisms that coordinate gene expression states are directly linked to mechanisms that regulate cell division in the developing nervous system. Epigenetic Regulation Of Neurogenesis By Citron Kinase Matthew J. Girgenti B.S., Fairfield University, 2002 M.S., Southern Connecticut State University, 2004 A Dissertation Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy at the University of Connecticut 2015 ii Copyright by Matthew J. -

Regulation of Titin-Based Cardiac Stiffness by Unfolded Domain Oxidation (Undox)
Regulation of titin-based cardiac stiffness by unfolded domain oxidation (UnDOx) Christine M. Loeschera,1, Martin Breitkreuzb,1, Yong Lia, Alexander Nickelc, Andreas Ungera, Alexander Dietld, Andreas Schmidte, Belal A. Mohamedf, Sebastian Kötterg, Joachim P. Schmitth, Marcus Krügere,i, Martina Krügerg, Karl Toischerf, Christoph Maackc, Lars I. Leichertj, Nazha Hamdanib, and Wolfgang A. Linkea,2 aInstitute of Physiology II, University of Munster, 48149 Munster, Germany; bInstitute of Physiology, Ruhr University Bochum, 44801 Bochum, Germany; cComprehensive Heart Failure Center Wuerzburg, University Clinic Wuerzburg, 97078 Wuerzburg, Germany; dDepartment of Internal Medicine II, University Hospital Regensburg, 93053 Regensburg, Germany; eInstitute for Genetics, University of Cologne, 50931 Cologne, Germany; fDepartment of Cardiology and Pneumology, University Medicine Goettingen, 37075 Goettingen, Germany; gDepartment of Cardiovascular Physiology, Heinrich Heine University, 40225 Düsseldorf, Germany; hDepartment of Pharmacology and Clinical Pharmacology, Heinrich Heine University, 40225 Düsseldorf, Germany; iCenter for Molecular Medicine and Excellence Cluster "Cellular Stress Responses in Aging-Associated Diseases" (CECAD), University of Cologne, 50931 Cologne, Germany; and jInstitute for Biochemistry and Pathobiochemistry, Ruhr University Bochum, 44801 Bochum, Germany Edited by Jonathan Seidman, Harvard University, Boston, MA, and approved August 12, 2020 (received for review March 14, 2020) The relationship between oxidative stress and -

Postmortem Changes in the Myofibrillar and Other Cytoskeletal Proteins in Muscle
BIOCHEMISTRY - IMPACT ON MEAT TENDERNESS Postmortem Changes in the Myofibrillar and Other C'oskeletal Proteins in Muscle RICHARD M. ROBSON*, ELISABETH HUFF-LONERGAN', FREDERICK C. PARRISH, JR., CHIUNG-YING HO, MARVIN H. STROMER, TED W. HUIATT, ROBERT M. BELLIN and SUZANNE W. SERNETT introduction filaments (titin), and integral Z-line region (a-actinin, Cap Z), as well as proteins of the intermediate filaments (desmin, The cytoskeleton of "typical" vertebrate cells contains paranemin, and synemin), Z-line periphery (filamin) and three protein filament systems, namely the -7-nm diameter costameres underlying the cell membrane (filamin, actin-containing microfilaments, the -1 0-nm diameter in- dystrophin, talin, and vinculin) are listed along with an esti- termediate filaments (IFs), and the -23-nm diameter tubu- mate of their abundance, approximate molecular weights, lin-containing microtubules (Robson, 1989, 1995; Robson and number of subunits per molecule. Because the myofibrils et al., 1991 ).The contractile myofibrils, which are by far the are the overwhelming components of the skeletal muscle cell major components of developed skeletal muscle cells and cytoskeleton, the approximate percentages of the cytoskel- are responsible for most of the desirable qualities of muscle eton listed for the myofibrillar proteins (e.g., myosin, actin, foods (Robson et al., 1981,1984, 1991 1, can be considered tropomyosin, a-actinin, etc.) also would represent their ap- the highly expanded corollary of the microfilament system proximate percentages of total myofibrillar protein. of non-muscle cells. The myofibrils, IFs, cell membrane skel- eton (complex protein-lattice subjacent to the sarcolemma), Some Important Characteristics, Possible and attachment sites connecting these elements will be con- Roles, and Postmortem Changes of Key sidered as comprising the muscle cell cytoskeleton in this Cytoskeletal Proteins review. -

Α-Actinin/Titin Interaction: a Dynamic and Mechanically Stable Cluster of Bonds in the Muscle Z-Disk
α-Actinin/titin interaction: A dynamic and mechanically stable cluster of bonds in the muscle Z-disk Marco Grisona, Ulrich Merkela, Julius Kostanb, Kristina Djinovic-Carugob,c, and Matthias Riefa,d,1 aPhysik Department E22, Technische Universität München, 85748 Garching, Germany; bDepartment of Structural and Computational Biology, Max F. Perutz Laboratories, University of Vienna, A-1030 Vienna, Austria; cDepartment of Biochemistry, Faculty of Chemistry and Chemical Technology, University of Ljubljana, SI-1000 Ljubljana, Slovenia; and dMunich Center for Integrated Protein Science, 81377 Munich, Germany Edited by James A. Spudich, Stanford University School of Medicine, Stanford, CA, and approved December 16, 2016 (received for review August 2, 2016) Stable anchoring of titin within the muscle Z-disk is essential for In humans, the isoforms of titin exhibit four to seven Z-repeats preserving muscle integrity during passive stretching. One of the (15, 16, 20). The structure of the EF3-4 hands complex with titin main candidates for anchoring titin in the Z-disk is the actin cross- Z-repeat 7 shows the bound Z-repeat in an α-helical confor- linker α-actinin. The calmodulin-like domain of α-actinin binds to mation (21). In solution assays, binding affinities of various the Z-repeats of titin. However, the mechanical and kinetic prop- Z-repeats to EF3-4 were determined to lie in the micromolar erties of this important interaction are still unknown. Here, we use range (22). Micromolar affinity points only to a moderately a dual-beam optical tweezers assay to study the mechanics of this stable interaction, the kinetics of which are unknown. -

Jadomycin B, an Aurora-B Kinase Inhibitor Discovered Through Virtual Screening
2386 Jadomycin B, an Aurora-B kinase inhibitor discovered through virtual screening Da-Hua Fu,1 Wei Jiang,1 Jian-Ting Zheng,2 Jadomycin B is a new Aurora-B kinase inhibitor worthy of Gui-Yu Zhao,1 Yan Li,1 Hong Yi,1 Zhuo-Rong Li,1 further investigation. [Mol Cancer Ther 2008;7(8):2386–93] Jian-Dong Jiang,1 Ke-Qian Yang,2 Yanchang Wang,3 and Shu-Yi Si1 Introduction In mammals, there are three Aurora kinases, Aurora-A, B, 1Institute of Medicinal Biotechnology, Peking Union Medical College & Chinese Academy of Medical Sciences, and 2Institute and C, which display >60% sequence identity (1). Although of Microbiology, Chinese Academy of Sciences, Beijing, China; the catalytic domains of the three kinases are highly and 3Department of Biomedical Sciences, College of Medicine, conserved, they show distinct subcellular localization and Florida State University, Tallahassee, Florida biological function (2, 3). Aurora-A, which is required for centrosome maturation and separation, localizes to centro- somes from early S phase to late M phase (4). Aurora-B is a Abstract component of chromosomal passenger complex, whose Aurora kinases have emerged as promising targets for function in mitosis has been extensively studied. In cancer therapy because of their critical role in mitosis. addition to Aurora-B, the chromosomal passenger complex These kinases are well-conserved in all eukaryotes, and contains Survivin, Borealin, and inner centromere protein IPL1 gene encodes the single Aurora kinase in budding (5–7). The chromosomal passenger complex associates yeast. In a virtual screening attempt, 22 compounds were with centromeric regions of chromosomes in the early identified from nearly 15,000 microbial natural products as stages of mitosis, but it translocates to microtubules after potential small-molecular inhibitors of human Aurora-B the onset of anaphase. -
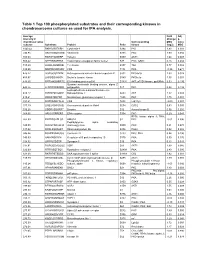
Table 1 Top 100 Phosphorylated Substrates and Their Corresponding Kinases in Chondrosarcoma Cultures As Used for IPA Analysis
Table 1 Top 100 phosphorylated substrates and their corresponding kinases in chondrosarcoma cultures as used for IPA analysis. Average Fold Adj intensity in Change p- chondrosarcoma Corresponding MSC value cultures Substrate Protein Psite kinase (log2) MSC 1043.42 RKKKVSSTKRH Cytohesin-1 S394 PKC 1.83 0.001 746.95 RKGYRSQRGHS Vitronectin S381 PKC 1.00 0.056 709.03 RARSTSLNERP Tuberin S939 AKT1 1.64 0.008 559.42 SPPRSSLRRSS Transcription elongation factor A-like1 S37 PKC; GSK3 0.18 0.684 515.29 LRRSLSRSMSQ Telethonin S157 Titin 0.77 0.082 510.00 MQPDNSSDSDY CD5 T434 PKA -0.35 0.671 476.27 GGRGGSRARNL Heterogeneous nuclear ribonucleoprotein K S302 PKCdelta 1.03 0.028 455.97 LKPGSSHRKTK Bruton's tyrosine kinase S180 PKCbeta 1.55 0.001 444.65 RRRMASMQRTG E1A binding protein p300 S1834 AKT; p70S6 kinase; pp90Rsk 0.53 0.195 Guanine nucleotide binding protein, alpha Z 440.26 HLRSESQRQRR polypeptide S27 PKC 0.88 0.199 6-phosphofructo-2-kinase/fructose-2,6- 424.12 RPRNYSVGSRP biphosphatase 2 S483 AKT 1.32 0.003 419.61 KKKIATRKPRF Metabotropic glutamate receptor 1 T695 PKC 1.75 0.001 391.21 DNSSDSDYDLH CD5 T453 Lck; Fyn -2.09 0.001 377.39 LRQLRSPRRAQ Ras associated protein Rab4 S204 CDC2 0.63 0.091 376.28 SSQRVSSYRRT Desmin S12 Aurora kinase B 0.56 0.255 369.05 ARIGGSRRERS EP4 receptor S354 PKC 0.29 0.543 RPS6 kinase alpha 3; PKA; 367.99 EPKRRSARLSA HMG14 S7 PKC -0.01 0.996 Peptidylglycine alpha amidating 349.08 SRKGYSRKGFD monooxygenase S930 PKC 0.21 0.678 347.92 RRRLSSLRAST Ribosomal protein S6 S236 PAK2 0.02 0.985 346.84 RSNPPSRKGSG Connexin -

Association of Titin and Myosin Heavy Chain in Developing Skeletal Muscle (Myogenesis/Cytoskeleton/Assembly in Vvo) W
Proc. Natl. Acad. Sci. USA Vol. 89, pp. 74%-7500, August 1992 Cell Biology Association of titin and myosin heavy chain in developing skeletal muscle (myogenesis/cytoskeleton/assembly in vvo) W. B. ISAACS*, I. S. KIM, A. STRUVE, AND A. B. FULTONt Department of Biochemistry, University of Iowa, Iowa City, IA 52242 Communicated by Sheldon Penman, May 22, 1992 ABSTRACT To understand molecular interactions that deficient medium (10). After labeling, cultures either were organize developing myoflbrils, we examined the biosynthesis extracted immediately or were chased by adding complete and interaction of titin and myosin heavy chain in cultures of medium supplemented with 2 mM unlabeled methionine and developing muscle. Use of pulse-labeling, immunoprecipita- incubating at 370C for various times before extraction. Ex- tion, and a reversible cross-linking procedure demonstrates tractions used 0.5% Triton X-100 in extraction buffer (100 that within minutes of synthesis, titin and myosin heavy chain mM KCI/10 mM Pipes, pH 6.8/300 mM sucrose/2 mM can be chemically cross-linked into very large, detergent- MgCI2/1 mM EGTA) containing protease inhibitors (1 mM resistant complexes retaining many features of intact myo- phenylmethylsulfonyl fluoride and 100 mM leupeptin; ref. tubes. These complexes, predominantly of titin and myosin, 11). occur very early in myofibrillogenesis as well as later. These Immunoprecipitation. Titin was precipitated by using a data suggest that synthesis and assembly oftitin and myosin are mouse monoclonal antibody (mAb), AMF-1, as described temporally and spatially coordinated in nascent myofibrils and (10). Muscle-specific myosin heavy chain (hereafter myosin) support the hypothesis that titin molecules help to organize was precipitated with mAb MF-20 (12), a gift ofD.