Supplementary Tables
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Sequences, Annotation and Single Nucleotide Polymorphism of The
Nova Southeastern University NSUWorks Biology Faculty Articles Department of Biological Sciences 6-2008 Sequences, Annotation and Single Nucleotide Polymorphism of the Major Histocompatibility Complex in the Domestic Cat Naoya Yuhki National Cancer Institute at Frederick James C. Mullikin National Human Genome Research Institute Thomas W. Beck National Cancer Institute at Frederick Robert M. Stephens National Cancer Institute at Frederick Stephen J. O'Brien National Cancer Institute at Frederick, [email protected] Follow this and additional works at: https://nsuworks.nova.edu/cnso_bio_facarticles Part of the Genetics and Genomics Commons, and the Zoology Commons NSUWorks Citation Yuhki, Naoya; James C. Mullikin; Thomas W. Beck; Robert M. Stephens; and Stephen J. O'Brien. 2008. "Sequences, Annotation and Single Nucleotide Polymorphism of the Major Histocompatibility Complex in the Domestic Cat." PLoS ONE 7, (3 e2674): 1-17. https://nsuworks.nova.edu/cnso_bio_facarticles/773 This Article is brought to you for free and open access by the Department of Biological Sciences at NSUWorks. It has been accepted for inclusion in Biology Faculty Articles by an authorized administrator of NSUWorks. For more information, please contact [email protected]. Sequences, Annotation and Single Nucleotide Polymorphism of the Major Histocompatibility Complex in the Domestic Cat Naoya Yuhki1*, James C. Mullikin2, Thomas Beck3, Robert Stephens4, Stephen J. O’Brien1 1 Laboratory of Genomic Diversity, National Cancer Institute at Frederick, Frederick, Maryland, -

2020 Program Book
PROGRAM BOOK Note that TAGC was cancelled and held online with a different schedule and program. This document serves as a record of the original program designed for the in-person meeting. April 22–26, 2020 Gaylord National Resort & Convention Center Metro Washington, DC TABLE OF CONTENTS About the GSA ........................................................................................................................................................ 3 Conference Organizers ...........................................................................................................................................4 General Information ...............................................................................................................................................7 Mobile App ....................................................................................................................................................7 Registration, Badges, and Pre-ordered T-shirts .............................................................................................7 Oral Presenters: Speaker Ready Room - Camellia 4.......................................................................................7 Poster Sessions and Exhibits - Prince George’s Exhibition Hall ......................................................................7 GSA Central - Booth 520 ................................................................................................................................8 Internet Access ..............................................................................................................................................8 -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -
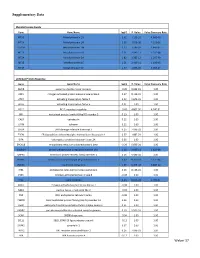
Weber 37 Supplementary Data
Supplementary Data Metallothionein Family Gene Gene Name logFC P. Value False Discovery Rate MT1G Metallothionein 1G 2.62 1.19E-10 4.66E-08 MT1H Metallothionein 1H 2.58 4.87E-08 7.12E-06 MT1M Metallothionein 1M 2.23 1.18E-07 1.44E-05 MT1X Metallothionein 1X 2.21 2.64E-11 1.25E-08 MT2A Metallothionein 2A 1.82 1.38E-12 1.26E-09 MT1E Metallothionein 1E 1.51 2.41E-11 1.18E-08 MT1F Metallothionein 1F 1.47 1.85E-09 4.09E-07 Unfolded Protein Response Gene Gene Name logFC P. Value False Discovery Rate AMFR autocrine motility factor receptor -0.20 9.94E-01 1.00 ASK1 mitogen-activated protein kinase kinase kinase 5 0.27 8.11E-01 1.00 ATF4 activating transcription factor 4 0.22 9.67E-01 1.00 ATF6 activating transcription factor 6 0.01 1.00 1.00 BCL2 BCL2, apoptosis regulator -0.43 4.80E-02 4.24E-01 BIP heat shock protein family A (Hsp70) member 5 0.13 1.00 1.00 CALR calreticulin 0.12 1.00 1.00 CANX calnexin 0.12 1.00 1.00 CHOP DNA damage inducible transcript 3 0.25 7.99E-01 1.00 EDEM ER degradation enhancing alpha-mannosidase like protein 1 0.37 1.88E-01 1.00 EIFA eukaryotic translation initiation factor 2A 0.06 1.00 1.00 ERO1LB endoplasmic reticulum oxidoreductase 1 beta -0.29 5.69E-01 1.00 GADD34 protein phosphatase 1 regulatory subunit 15A 1.18 3.66E-11 1.62E-08 GRP94 heat shock protein 90 beta family member 1 0.16 9.98E-01 1.00 HSPH1 heat shock protein family H (Hsp110) member 1 0.62 9.70E-06 5.12E-04 INSIG1 insulin induced gene 1 1.69 8.47E-18 3.88E-14 IRE1 endoplasmic reticulum to nucleus signaling 1 0.26 8.13E-01 1.00 JNK1 mitogen-activated -

Supplementary Material
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) J Neurol Neurosurg Psychiatry Page 1 / 45 SUPPLEMENTARY MATERIAL Appendix A1: Neuropsychological protocol. Appendix A2: Description of the four cases at the transitional stage. Table A1: Clinical status and center proportion in each batch. Table A2: Complete output from EdgeR. Table A3: List of the putative target genes. Table A4: Complete output from DIANA-miRPath v.3. Table A5: Comparison of studies investigating miRNAs from brain samples. Figure A1: Stratified nested cross-validation. Figure A2: Expression heatmap of miRNA signature. Figure A3: Bootstrapped ROC AUC scores. Figure A4: ROC AUC scores with 100 different fold splits. Figure A5: Presymptomatic subjects probability scores. Figure A6: Heatmap of the level of enrichment in KEGG pathways. Kmetzsch V, et al. J Neurol Neurosurg Psychiatry 2021; 92:485–493. doi: 10.1136/jnnp-2020-324647 BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) J Neurol Neurosurg Psychiatry Appendix A1. Neuropsychological protocol The PREV-DEMALS cognitive evaluation included standardized neuropsychological tests to investigate all cognitive domains, and in particular frontal lobe functions. The scores were provided previously (Bertrand et al., 2018). Briefly, global cognitive efficiency was evaluated by means of Mini-Mental State Examination (MMSE) and Mattis Dementia Rating Scale (MDRS). Frontal executive functions were assessed with Frontal Assessment Battery (FAB), forward and backward digit spans, Trail Making Test part A and B (TMT-A and TMT-B), Wisconsin Card Sorting Test (WCST), and Symbol-Digit Modalities test. -

Supplement 1A Steffensen Et
Liver Wild-type Knockout C T C T 1 2 4 7 8 9 1 2 4 5 7 9 1 1 1 1 1 1 C C C T T T C C C T T T W W W W W W K K K K K K IMAGE:793166 RIKEN cDNA 6720463E02 gene IMAGE:1447421 ESTs, Weakly similar to ZF37 MOUSE ZINC FINGER PROTEIN 37 [M.musculus] IMAGE:934291 RIKEN cDNA 2810418N01 gene IMAGE:1247525 small EDRK-rich f2actor IMAGE:1449402 expressed sequence AW321064 IMAGE:1279847 ESTs IMAGE:518737 expressed sequence AW049941 IMAGE:860231 a disintegrin and metalloproteinase domain 17 IMAGE:642836 CD86 antigen IMAGE:1003885 phosphoribosyl pyrophosphate sy1nthetase IMAGE:524862 RIKEN cDNA 5730469D23 gene IMAGE:1264473 protein inhibitor of activat1ed STAT IMAGE:847035 RIKEN cDNA 4833422F06 gene IMAGE:374550 requiem IMAGE:976520 nuclear receptor coact4ivator IMAGE:1264311 Unknown IMAGE:976735 expressed sequence AI987692 IMAGE:976659 cathepsLin IMAGE:1477580 RIKEN cDNA 1600010J02 gene IMAGE:1277168 ribosomal protein, large, P1 IMAGE:524842 RIKEN cDNA 0710008D09 gene IMAGE:373019 split hand/foot delete1d gene IMAGE:404428 expressed sequence AI413851 IMAGE:619810 RIKEN cDNA 1700003F10 gene IMAGE:1749558 caspase 3, apoptosis related cysteine protease IMAGE:718718 RIKEN cDNA 2810003F23 gene IMAGE:819789 Unknown IMAGE:524474 ATP-binding cassette, sub-family A ABC1, member IMAGE:804950 Mus musculus, Similar to ribosomal protein S20, clone MGC:6876 IMAGE:2651405, mRNA, complete cds IMAGE:806143 gap junction membrane channel prot2ein beta IMAGE:1745887 expressed sequence AI836376 IMAGE:779426 RIKEN cDNA 5230400G24 gene IMAGE:1125615 Unknown IMAGE:535025 DNA -

Appendix 2. Significantly Differentially Regulated Genes in Term Compared with Second Trimester Amniotic Fluid Supernatant
Appendix 2. Significantly Differentially Regulated Genes in Term Compared With Second Trimester Amniotic Fluid Supernatant Fold Change in term vs second trimester Amniotic Affymetrix Duplicate Fluid Probe ID probes Symbol Entrez Gene Name 1019.9 217059_at D MUC7 mucin 7, secreted 424.5 211735_x_at D SFTPC surfactant protein C 416.2 206835_at STATH statherin 363.4 214387_x_at D SFTPC surfactant protein C 295.5 205982_x_at D SFTPC surfactant protein C 288.7 1553454_at RPTN repetin solute carrier family 34 (sodium 251.3 204124_at SLC34A2 phosphate), member 2 238.9 206786_at HTN3 histatin 3 161.5 220191_at GKN1 gastrokine 1 152.7 223678_s_at D SFTPA2 surfactant protein A2 130.9 207430_s_at D MSMB microseminoprotein, beta- 99.0 214199_at SFTPD surfactant protein D major histocompatibility complex, class II, 96.5 210982_s_at D HLA-DRA DR alpha 96.5 221133_s_at D CLDN18 claudin 18 94.4 238222_at GKN2 gastrokine 2 93.7 1557961_s_at D LOC100127983 uncharacterized LOC100127983 93.1 229584_at LRRK2 leucine-rich repeat kinase 2 HOXD cluster antisense RNA 1 (non- 88.6 242042_s_at D HOXD-AS1 protein coding) 86.0 205569_at LAMP3 lysosomal-associated membrane protein 3 85.4 232698_at BPIFB2 BPI fold containing family B, member 2 84.4 205979_at SCGB2A1 secretoglobin, family 2A, member 1 84.3 230469_at RTKN2 rhotekin 2 82.2 204130_at HSD11B2 hydroxysteroid (11-beta) dehydrogenase 2 81.9 222242_s_at KLK5 kallikrein-related peptidase 5 77.0 237281_at AKAP14 A kinase (PRKA) anchor protein 14 76.7 1553602_at MUCL1 mucin-like 1 76.3 216359_at D MUC7 mucin 7, -

Rats and Axolotls Share a Common Molecular Signature After Spinal Cord Injury Enriched in Collagen-1
Rats and axolotls share a common molecular signature after spinal cord injury enriched in collagen-1 Athanasios Didangelos1, Katalin Bartus1, Jure Tica1, Bernd Roschitzki2, Elizabeth J. Bradbury1 1Wolfson CARD King’s College London, United Kingdom. 2Centre for functional Genomics, ETH Zurich, Switzerland. Running title: spinal cord injury in rats and axolotls Correspondence: A Didangelos: [email protected] SUPPLEMENTAL FIGURES AND LEGENDS Supplemental Fig. 1: Rat 7 days microarray differentially regulated transcripts. A-B: Protein-protein interaction networks of upregulated (A) and downregulated (B) transcripts identified by microarray gene expression profiling of rat SCI (4 sham versus 4 injured spinal cord samples) 7 days post-injury. Microarray expression data and experimental information is publicly available online (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE45006) and is also summarised in Supplemental Table 1. Protein-protein interaction networks were performed in StringDB using the full range of protein interaction scores (0.15 – 0.99) to capture maximum evidence of proteins’ interactions. Networks were then further analysed for betweeness centrality and gene ontology (GO) annotations (BinGO) in Cytoscape. Node colour indicates betweeness centrality while edge colour and thickness indicate interaction score based on predicted functional links between nodes (green: low values; red: high values). C-D: The top 10 upregulated (C) or downregulated (D) transcripts sorted by betweeness centrality score in protein-protein interaction networks shown in A & B. E-F: Biological process GO analysis was performed on networks of upregulated and downregulated genes using BinGO in Cytoscape. Graphs indicate the 20 most significant GO categories and the number of genes in each category. -
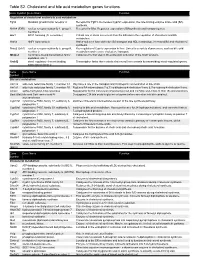
Figures Intermediary Ontogeny.Pptx
Table S2. Cholesterol and bile acid metabolism genes functions. Gene Symbol Gene Name Function Regulation of cholesterol and/or bile acid metabolism Fgfr4 fibroblast growth factor receptor 4 Receptor for Fgf15. Decreases Cyp7a1 expression, the rate-limiting enzyme in bile acid (BA) synthesis. Nr1h4 (FXR) nuclear receptor subfamily 1, group H, Receptor for BAs. Regulates expression of BAsynthesis and transport genes. member 4 Arv1 ARV1 homolog (S. cerevisiae) Critical role in sterol movement from the ER and in the regulation of cholesterol and BA metabolism. Hnf1a HNF1 homeobox A Hnf1a-null mice have defective BA transport and HDL metabolism, increased BA and cholesterol synthesis. Nr5a2 (Lrh1) nuclear receptor subfamily 5, group A, Key regulator of Cyp7a expression in liver. Linked to a variety of processes, such as bile acid member 2 metabolism and reverse cholesterol transport. Mbtps1 membrane-bound transcription factor Catalyzes the first step in the proteolytic activation of the Srebf proteins. peptidase, site 1 Srebf2 sterol regulatory element binding Transcription factor that controls cholesterol homeostasis by transcribing sterol-regulated genes. transcription factor 2 Gene Gene Name Function Symbol Bile acid metabolism Akr1c6 aldo-keto reductase family 1, member C1 May have a role in the transport and intrahepatic concentration of bile acids. Akr1d1 aldo-keto reductase family 1, member D1 Reduces BA intermediates 7-α,12-α-dihydroxy-4-cholesten-3-one & 7-α-hydroxy-4-cholesten-3-one. Amacr alpha-methylacyl-CoA racemase Responsible for the conversion of pristanoyl-CoA and C27-bile acyl-CoAs to their (S)-stereoisomers. Baat (Bat) bile acid CoA: amino acid N- Conjugates C24 bile acids to glycine or taurine before excretion into bile canaliculi. -

Anti-C6orf136 Product Datasheet Polyclonal Antibody
Anti-C6orf136 Product Datasheet Polyclonal Antibody PRODUCT SPECIFICATION Product Name Anti-C6orf136 Product Number HPA046804 Gene Description chromosome 6 open reading frame 136 Clonality Polyclonal Isotype IgG Host Rabbit Antigen Sequence Recombinant Protein Epitope Signature Tag (PrEST) antigen sequence: EGPGPELHSGCLDGLRSLFEGPPCPYPGAWIPFQVPGTAHPSPATPSGDP SMEEHLSVMYERLRQELPKLFLQSHDYSLYSLDV Purification Method Affinity purified using the PrEST antigen as affinity ligand Verified Species Human Reactivity Recommended IHC (Immunohistochemistry) Applications - Antibody dilution: 1:20 - 1:50 - Retrieval method: HIER pH6 Characterization Data Available at atlasantibodies.com/products/HPA046804 Buffer 40% glycerol and PBS (pH 7.2). 0.02% sodium azide is added as preservative. Concentration Lot dependent Storage Store at +4°C for short term storage. Long time storage is recommended at -20°C. Notes Gently mix before use. Optimal concentrations and conditions for each application should be determined by the user. For protocols, additional product information, such as images and references, see atlasantibodies.com. Product of Sweden. For research use only. Not intended for pharmaceutical development, diagnostic, therapeutic or any in vivo use. No products from Atlas Antibodies may be resold, modified for resale or used to manufacture commercial products without prior written approval from Atlas Antibodies AB. Warranty: The products supplied by Atlas Antibodies are warranted to meet stated product specifications and to conform to label descriptions when used and stored properly. Unless otherwise stated, this warranty is limited to one year from date of sales for products used, handled and stored according to Atlas Antibodies AB's instructions. Atlas Antibodies AB's sole liability is limited to replacement of the product or refund of the purchase price. -

Download Special Issue
BioMed Research International Novel Bioinformatics Approaches for Analysis of High-Throughput Biological Data Guest Editors: Julia Tzu-Ya Weng, Li-Ching Wu, Wen-Chi Chang, Tzu-Hao Chang, Tatsuya Akutsu, and Tzong-Yi Lee Novel Bioinformatics Approaches for Analysis of High-Throughput Biological Data BioMed Research International Novel Bioinformatics Approaches for Analysis of High-Throughput Biological Data Guest Editors: Julia Tzu-Ya Weng, Li-Ching Wu, Wen-Chi Chang, Tzu-Hao Chang, Tatsuya Akutsu, and Tzong-Yi Lee Copyright © 2014 Hindawi Publishing Corporation. All rights reserved. This is a special issue published in “BioMed Research International.” All articles are open access articles distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Contents Novel Bioinformatics Approaches for Analysis of High-Throughput Biological Data,JuliaTzu-YaWeng, Li-Ching Wu, Wen-Chi Chang, Tzu-Hao Chang, Tatsuya Akutsu, and Tzong-Yi Lee Volume2014,ArticleID814092,3pages Evolution of Network Biomarkers from Early to Late Stage Bladder Cancer Samples,Yung-HaoWong, Cheng-Wei Li, and Bor-Sen Chen Volume 2014, Article ID 159078, 23 pages MicroRNA Expression Profiling Altered by Variant Dosage of Radiation Exposure,Kuei-FangLee, Yi-Cheng Chen, Paul Wei-Che Hsu, Ingrid Y. Liu, and Lawrence Shih-Hsin Wu Volume2014,ArticleID456323,10pages EXIA2: Web Server of Accurate and Rapid Protein Catalytic Residue Prediction, Chih-Hao Lu, Chin-Sheng