Supplement 1A Steffensen Et
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplementary Data
Supplementary Data for Quantitative Changes in the Mitochondrial Proteome from Subjects with Mild Cognitive Impairment, Early Stage and Late Stage Alzheimer’s disease Table 1 - 112 unique, non-redundant proteins identified and quantified in at least two of the three analytical replicates for all three disease stages. Table 2 - MCI mitochondrial samples, Protein Summary Table 3 - MCI mitochondrial samples, Experiment 1 Table 4 - MCI mitochondrial samples, Experiment 2 Table 5 - MCI mitochondrial samples, Experiment 3 Table 6 - EAD Mitochondrial Study, Protein Summary Table 7 - EAD Mitochondrial Study, Experiment 1 Table 8 - EAD Mitochondrial Study, Experiment 2 Table 9 - EAD Mitochondrial Study, Experiment 3 Table 10 - LAD Mitochondrial Study, Protein Summary Table 11 - LAD Mitochondrial Study, Experiment 1 Table 12 - LAD Mitochondrial Study, Experiment 2 Table 13 - LAD Mitochondrial Study, Experiment 3 Supplemental Table 1. 112 unique, non-redundant proteins identified and quantified in at least two of the three analytical replicates for all three disease stages. Description Data MCI EAD LAD AATM_HUMAN (P00505) Aspartate aminotransferase, mitochondrial precursor (EC Mean 1.43 1.70 1.31 2.6.1.1) (Transaminase A) (Glutamate oxaloacetate transaminase 2) [MASS=47475] SEM 0.07 0.09 0.09 Count 3.00 3.00 3.00 ACON_HUMAN (Q99798) Aconitate hydratase, mitochondrial precursor (EC 4.2.1.3) Mean 1.24 1.61 1.19 (Citrate hydro-lyase) (Aconitase) [MASS=85425] SEM 0.05 0.17 0.18 Count 3.00 2.00 3.00 ACPM_HUMAN (O14561) Acyl carrier protein, mitochondrial -

Table 2. Significant
Table 2. Significant (Q < 0.05 and |d | > 0.5) transcripts from the meta-analysis Gene Chr Mb Gene Name Affy ProbeSet cDNA_IDs d HAP/LAP d HAP/LAP d d IS Average d Ztest P values Q-value Symbol ID (study #5) 1 2 STS B2m 2 122 beta-2 microglobulin 1452428_a_at AI848245 1.75334941 4 3.2 4 3.2316485 1.07398E-09 5.69E-08 Man2b1 8 84.4 mannosidase 2, alpha B1 1416340_a_at H4049B01 3.75722111 3.87309653 2.1 1.6 2.84852656 5.32443E-07 1.58E-05 1110032A03Rik 9 50.9 RIKEN cDNA 1110032A03 gene 1417211_a_at H4035E05 4 1.66015788 4 1.7 2.82772795 2.94266E-05 0.000527 NA 9 48.5 --- 1456111_at 3.43701477 1.85785922 4 2 2.8237185 9.97969E-08 3.48E-06 Scn4b 9 45.3 Sodium channel, type IV, beta 1434008_at AI844796 3.79536664 1.63774235 3.3 2.3 2.75319499 1.48057E-08 6.21E-07 polypeptide Gadd45gip1 8 84.1 RIKEN cDNA 2310040G17 gene 1417619_at 4 3.38875643 1.4 2 2.69163229 8.84279E-06 0.0001904 BC056474 15 12.1 Mus musculus cDNA clone 1424117_at H3030A06 3.95752801 2.42838452 1.9 2.2 2.62132809 1.3344E-08 5.66E-07 MGC:67360 IMAGE:6823629, complete cds NA 4 153 guanine nucleotide binding protein, 1454696_at -3.46081884 -4 -1.3 -1.6 -2.6026947 8.58458E-05 0.0012617 beta 1 Gnb1 4 153 guanine nucleotide binding protein, 1417432_a_at H3094D02 -3.13334396 -4 -1.6 -1.7 -2.5946297 1.04542E-05 0.0002202 beta 1 Gadd45gip1 8 84.1 RAD23a homolog (S. -

An Anticomplement Agent That Homes to the Damaged Brain and Promotes Recovery After Traumatic Brain Injury in Mice
An anticomplement agent that homes to the damaged brain and promotes recovery after traumatic brain injury in mice Marieta M. Rusevaa,1,2, Valeria Ramagliab,1, B. Paul Morgana, and Claire L. Harrisa,3 aInstitute of Infection and Immunity, School of Medicine, Cardiff University, Cardiff CF14 4XN, United Kingdom; and bDepartment of Genome Analysis, Academic Medical Center, Amsterdam 1105 AZ, The Netherlands Edited by Douglas T. Fearon, Cornell University, Cambridge, United Kingdom, and approved September 29, 2015 (received for review July 15, 2015) Activation of complement is a key determinant of neuropathology to rapidly and specifically inhibit MAC at sites of complement and disability after traumatic brain injury (TBI), and inhibition is activation, and test its therapeutic potential in experimental TBI. neuroprotective. However, systemic complement is essential to The construct, termed CD59-2a-CRIg, comprises CD59a linked fight infections, a critical complication of TBI. We describe a to CRIg via the murine IgG2a hinge. CD59a prevents assembly targeted complement inhibitor, comprising complement receptor of MAC in cell membranes (16), whereas CRIg binds C3b/iC3b of the Ig superfamily (CRIg) fused with complement regulator CD59a, deposited at sites of complement activation (17). The IgG2a designed to inhibit membrane attack complex (MAC) assembly at hinge promotes dimerization to increase ligand avidity. CD59- sites of C3b/iC3b deposition. CRIg and CD59a were linked via the 2a-CRIg protected in the TBI model, demonstrating that site- IgG2a hinge, yielding CD59-2a-CRIg dimer with increased iC3b/C3b targeted anti-MAC therapeutics may be effective in prevention binding avidity and MAC inhibitory activity. CD59-2a-CRIg inhibited of secondary neuropathology and improve neurologic recovery MAC formation and prevented complement-mediated lysis in vitro. -

Peroxisomal Functions in the Lung and Their Role in the Pathogenesis of Lung Diseases
Aus dem Institut für Anatomie und Zellbiologie der Justus-Liebig-Universität Gießen Leiter: Prof. Dr. Eveline Baumgart-Vogt Peroxisomal functions in the lung and their role in the pathogenesis of lung diseases Habilitationsschrift zur Erlangung der Venia legendi des Fachbereichs Medizin der Justus-Liebig-Universität Gießen vorgelegt von Srikanth Karnati Gießen 2018 Die nachfolgende Arbeit nimmt Bezug auf folgende Originalarbeiten: 1. Karnati S*, Graulich T, Oruqaj G, Pfreimer S, Seimetz M, Stamme C, Mariani TJ, Weissmann N, Mühlfeld C, Baumgart-Vogt E (2016). Postnatal development of the secretory cells of the distal airways, the bronchiolar club cells in the mouse lung: stereological and molecular biological studies. Cell and Tissue Research. Jun;364(3):543- 57. 2. Karnati S, Baumgart-Vogt E (2009) PeroXisomes in airway epithelia and future prospects of these organelles for pulmonary cell biology. Histochem Cell Biol. Apr: 131(4):447-54. 3. Karnati S, Lüers G, Pfreimer S and Baumgart-Vogt E (2013) Manganese SuperoXide dismutase 2 (MnSOD) is localized to mitochondria but not in peroXisomes. Histochemistry and Cell Biology, Aug:140(2):105-17 4. Karnati S, Palaniswamy S, Alam MR, Oruqaj G, Stamme C, Baumgart-Vogt E (2015) C22- bronchial and T7-alveolar epithelial cell lines of the immortomouse are eXcellent murine cell culture model systems to study pulmonary peroXisome biology and metabolism. Histochemistry and Cell Biology Mar;145(3):287-304. 5. Oruqaj G§, Karnati S§, Vijayan V, Kotarkonda LK, Boateng E, Zhang W, Ruppert C, Günther A, Shi W, Baumgart-Vogt E (2015) Compromised peroXisomes in idiopathic pulmonary fibrosis, a vicious cycle inducing a higher fibrotic response via TGF-β signaling. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -
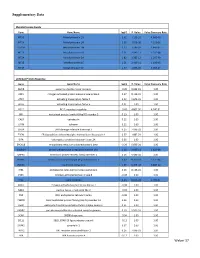
Weber 37 Supplementary Data
Supplementary Data Metallothionein Family Gene Gene Name logFC P. Value False Discovery Rate MT1G Metallothionein 1G 2.62 1.19E-10 4.66E-08 MT1H Metallothionein 1H 2.58 4.87E-08 7.12E-06 MT1M Metallothionein 1M 2.23 1.18E-07 1.44E-05 MT1X Metallothionein 1X 2.21 2.64E-11 1.25E-08 MT2A Metallothionein 2A 1.82 1.38E-12 1.26E-09 MT1E Metallothionein 1E 1.51 2.41E-11 1.18E-08 MT1F Metallothionein 1F 1.47 1.85E-09 4.09E-07 Unfolded Protein Response Gene Gene Name logFC P. Value False Discovery Rate AMFR autocrine motility factor receptor -0.20 9.94E-01 1.00 ASK1 mitogen-activated protein kinase kinase kinase 5 0.27 8.11E-01 1.00 ATF4 activating transcription factor 4 0.22 9.67E-01 1.00 ATF6 activating transcription factor 6 0.01 1.00 1.00 BCL2 BCL2, apoptosis regulator -0.43 4.80E-02 4.24E-01 BIP heat shock protein family A (Hsp70) member 5 0.13 1.00 1.00 CALR calreticulin 0.12 1.00 1.00 CANX calnexin 0.12 1.00 1.00 CHOP DNA damage inducible transcript 3 0.25 7.99E-01 1.00 EDEM ER degradation enhancing alpha-mannosidase like protein 1 0.37 1.88E-01 1.00 EIFA eukaryotic translation initiation factor 2A 0.06 1.00 1.00 ERO1LB endoplasmic reticulum oxidoreductase 1 beta -0.29 5.69E-01 1.00 GADD34 protein phosphatase 1 regulatory subunit 15A 1.18 3.66E-11 1.62E-08 GRP94 heat shock protein 90 beta family member 1 0.16 9.98E-01 1.00 HSPH1 heat shock protein family H (Hsp110) member 1 0.62 9.70E-06 5.12E-04 INSIG1 insulin induced gene 1 1.69 8.47E-18 3.88E-14 IRE1 endoplasmic reticulum to nucleus signaling 1 0.26 8.13E-01 1.00 JNK1 mitogen-activated -

Appendix 2. Significantly Differentially Regulated Genes in Term Compared with Second Trimester Amniotic Fluid Supernatant
Appendix 2. Significantly Differentially Regulated Genes in Term Compared With Second Trimester Amniotic Fluid Supernatant Fold Change in term vs second trimester Amniotic Affymetrix Duplicate Fluid Probe ID probes Symbol Entrez Gene Name 1019.9 217059_at D MUC7 mucin 7, secreted 424.5 211735_x_at D SFTPC surfactant protein C 416.2 206835_at STATH statherin 363.4 214387_x_at D SFTPC surfactant protein C 295.5 205982_x_at D SFTPC surfactant protein C 288.7 1553454_at RPTN repetin solute carrier family 34 (sodium 251.3 204124_at SLC34A2 phosphate), member 2 238.9 206786_at HTN3 histatin 3 161.5 220191_at GKN1 gastrokine 1 152.7 223678_s_at D SFTPA2 surfactant protein A2 130.9 207430_s_at D MSMB microseminoprotein, beta- 99.0 214199_at SFTPD surfactant protein D major histocompatibility complex, class II, 96.5 210982_s_at D HLA-DRA DR alpha 96.5 221133_s_at D CLDN18 claudin 18 94.4 238222_at GKN2 gastrokine 2 93.7 1557961_s_at D LOC100127983 uncharacterized LOC100127983 93.1 229584_at LRRK2 leucine-rich repeat kinase 2 HOXD cluster antisense RNA 1 (non- 88.6 242042_s_at D HOXD-AS1 protein coding) 86.0 205569_at LAMP3 lysosomal-associated membrane protein 3 85.4 232698_at BPIFB2 BPI fold containing family B, member 2 84.4 205979_at SCGB2A1 secretoglobin, family 2A, member 1 84.3 230469_at RTKN2 rhotekin 2 82.2 204130_at HSD11B2 hydroxysteroid (11-beta) dehydrogenase 2 81.9 222242_s_at KLK5 kallikrein-related peptidase 5 77.0 237281_at AKAP14 A kinase (PRKA) anchor protein 14 76.7 1553602_at MUCL1 mucin-like 1 76.3 216359_at D MUC7 mucin 7, -

Focused Transcription from the Human CR2/CD21 Core Promoter Is Regulated by Synergistic Activity of TATA and Initiator Elements in Mature B Cells
Cellular & Molecular Immunology (2016) 13, 119–131 ß 2015 CSI and USTC. All rights reserved 1672-7681/15 $32.00 www.nature.com/cmi RESEARCH ARTICLE Focused transcription from the human CR2/CD21 core promoter is regulated by synergistic activity of TATA and Initiator elements in mature B cells Rhonda L Taylor1,2, Mark N Cruickshank3, Mahdad Karimi2, Han Leng Ng1, Elizabeth Quail2, Kenneth M Kaufman4,5, John B Harley4,5, Lawrence J Abraham1, Betty P Tsao6, Susan A Boackle7 and Daniela Ulgiati1 Complement receptor 2 (CR2/CD21) is predominantly expressed on the surface of mature B cells where it forms part of a coreceptor complex that functions, in part, to modulate B-cell receptor signal strength. CR2/CD21 expression is tightly regulated throughout B-cell development such that CR2/CD21 cannot be detected on pre-B or terminally differentiated plasma cells. CR2/CD21 expression is upregulated at B-cell maturation and can be induced by IL-4 and CD40 signaling pathways. We have previously characterized elements in the proximal promoter and first intron of CR2/CD21 that are involved in regulating basal and tissue-specific expression. We now extend these analyses to the CR2/CD21 core promoter. We show that in mature B cells, CR2/CD21 transcription proceeds from a focused TSS regulated by a non-consensus TATA box, an initiator element and a downstream promoter element. Furthermore, occupancy of the general transcriptional machinery in pre-B versus mature B-cell lines correlate with CR2/CD21 expression level and indicate that promoter accessibility must switch from inactive to active during the transitional B-cell window. -

Structural Insight on the Recognition of Surface-Bound Opsonins by the Integrin I Domain of Complement Receptor 3
Structural insight on the recognition of surface-bound opsonins by the integrin I domain of complement receptor 3 Goran Bajica, Laure Yatimea, Robert B. Simb, Thomas Vorup-Jensenc,1, and Gregers R. Andersena,1 Departments of aMolecular Biology and Genetics and cBiomedicine, Aarhus University, DK-8000 Aarhus, Denmark; and bDepartment of Pharmacology, University of Oxford, Oxford OX1 3QT, United Kingdom Edited by Douglas T. Fearon, University of Cambridge School of Clinical Medicine, Cambridge, United Kingdom, and approved August 28, 2013 (received for review June 13, 2013) Complement receptors (CRs), expressed notably on myeloid and degranulation, and changes in leukocyte cytokine production (2, lymphoid cells, play an essential function in the elimination of 5–7). CR3, and to a lesser degree CR4, are essential for the complement-opsonized pathogens and apoptotic/necrotic cells. In phagocytosis of complement-opsonized particles or complexes addition, these receptors are crucial for the cross-talk between the (6, 8, 9). Complement-opsonized immune complexes are cap- innate andadaptive branches ofthe immune system. CR3 (also known tured in the lymph nodes by CR3-positive subcapsular sinus as Mac-1, integrin α β , or CD11b/CD18) is expressed on all macro- macrophages (SSMs) and conveyed directly to naïve B cells or M 2 γ phages and recognizes iC3b on complement-opsonized objects, en- through follicular dendritic cells (10) using CR1, CR2, and Fc abling their phagocytosis. We demonstrate that the C3d moiety of receptors for antigen capture (11, 12). Hence, antigen-presenting iC3b harbors the binding site for the CR3 αI domain, and our structure cells such as SSMs may act as antigen storage and provide B of the C3d:αI domain complex rationalizes the CR3 selectivity for iC3b. -

Genetic Defects in B-Cell Development and Their Clinical Consequences H Abolhassani,1,2 N Parvaneh,1 N Rezaei,1 L Hammarström,2 a Aghamohammadi1
REVIEWS Genetic Defects in B-Cell Development and Their Clinical Consequences H Abolhassani,1,2 N Parvaneh,1 N Rezaei,1 L Hammarström,2 A Aghamohammadi1 1Research Center for Immunodeficiencies, Pediatrics Center of Excellence, Children’s Medical Center, Tehran University of Medical Sciences, Tehran, Iran 2Division of Clinical Immunology, Department of Laboratory Medicine, Karolinska Institutet at Karolinska University Hospital Huddinge, Stockholm, Sweden n Abstract Expression of selected genes in hematopoietic stem cells has been identified as a regulator of differentiation of B cells in the liver and bone marrow. Moreover, naïve B cells expressing surface immunoglobulin need other types of genes for antigen-dependent development in secondary lymphoid organs. Many advanced molecular mechanisms underlying primary antibody deficiencies in humans have been described. We provide an overview of the mutations in genes known to be involved in B-cell development and their clinical consequences. Key words: Genetic disorder. B-cell development. Primary antibody deficiencies. Clinical phenotypes. n Resumen Se ha identificado la expresión de genes seleccionados en las células pluripotenciales de médula ósea como reguladores de la diferenciación de las células B en el hígado y en médula ósea. Sin embargo, las células B naïve que expresan inmunoglubulinas de superficie, necesitan otros tipos de genes para su desarrollo en los órganos linfoides secundarios dependienteS de antígeno. Se han descrito muchos mecanismos moleculares avanzados que subrayan las inmunodeficiencias en humanos y esta revisión constituye una visión general de la mutación en todos los genes conocidos involucrados en el desarrollo de las células B y sus consecuencias clínicas. Palabras clave: Alteraciones genéticas. Desarrollo de las células B. -

Hereditary Hearing Impairment with Cutaneous Abnormalities
G C A T T A C G G C A T genes Review Hereditary Hearing Impairment with Cutaneous Abnormalities Tung-Lin Lee 1 , Pei-Hsuan Lin 2,3, Pei-Lung Chen 3,4,5,6 , Jin-Bon Hong 4,7,* and Chen-Chi Wu 2,3,5,8,* 1 Department of Medical Education, National Taiwan University Hospital, Taipei City 100, Taiwan; [email protected] 2 Department of Otolaryngology, National Taiwan University Hospital, Taipei 11556, Taiwan; [email protected] 3 Graduate Institute of Clinical Medicine, National Taiwan University College of Medicine, Taipei City 100, Taiwan; [email protected] 4 Graduate Institute of Medical Genomics and Proteomics, National Taiwan University College of Medicine, Taipei City 100, Taiwan 5 Department of Medical Genetics, National Taiwan University Hospital, Taipei 10041, Taiwan 6 Department of Internal Medicine, National Taiwan University Hospital, Taipei 10041, Taiwan 7 Department of Dermatology, National Taiwan University Hospital, Taipei City 100, Taiwan 8 Department of Medical Research, National Taiwan University Biomedical Park Hospital, Hsinchu City 300, Taiwan * Correspondence: [email protected] (J.-B.H.); [email protected] (C.-C.W.) Abstract: Syndromic hereditary hearing impairment (HHI) is a clinically and etiologically diverse condition that has a profound influence on affected individuals and their families. As cutaneous findings are more apparent than hearing-related symptoms to clinicians and, more importantly, to caregivers of affected infants and young individuals, establishing a correlation map of skin manifestations and their underlying genetic causes is key to early identification and diagnosis of syndromic HHI. In this article, we performed a comprehensive PubMed database search on syndromic HHI with cutaneous abnormalities, and reviewed a total of 260 relevant publications. -

Complement Receptor 1 Therapeutics for Prevention of Immune Hemolysis
Review: complement receptor 1 therapeutics for prevention of immune hemolysis K.YAZDANBAKHSH The complement system plays a crucial role in fighting infections biological activities, it has to be activated. Activation and is an important link between the innate and adaptive immune occurs in a sequence that involves proteolytic cleavage responses. However, inappropriate complement activation can cause tissue damage, and it underlies the pathology of many of the complement components, resulting in the diseases. In the transfusion medicine setting, complement release of active biological mediators and the assembly sensitization of RBCs can lead to both intravascular and of active enzyme molecules that result in cleavage of extravascular destruction. Moreover, complement deficiencies are 1 associated with autoimmune disorders, including autoimmune the next downstream complement component. hemolytic anemia (AIHA). Complement receptor 1 (CR1) is a large Depending on the nature of the activators, three single-pass glycoprotein that is expressed on a variety of cell types complement activation pathways have been described: in blood, including RBCs and immune cells. Among its multiple the antibody-dependent classical pathway and the functions is its ability to inhibit complement activation. Furthermore, gene knockout studies in mice implicate a role for antibody-independent alternative and lectin pathways CR1 (along with the alternatively spliced gene product CR2) in (Fig. 1).1 Common to all three pathways are two prevention of autoimmunity. This review discusses the possibility critical steps: the assembly of the C3 convertase that the CR1 protein may be manipulated to prevent and treat AIHA. In addition, it will be shown in an in vivo mouse model of enzymes and the activation of C5 convertases.