Figures Intermediary Ontogeny.Pptx
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Characterization of the Ergosterol Biosynthesis Pathway in Ceratocystidaceae
Journal of Fungi Article Characterization of the Ergosterol Biosynthesis Pathway in Ceratocystidaceae Mohammad Sayari 1,2,*, Magrieta A. van der Nest 1,3, Emma T. Steenkamp 1, Saleh Rahimlou 4 , Almuth Hammerbacher 1 and Brenda D. Wingfield 1 1 Department of Biochemistry, Genetics and Microbiology, Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, Pretoria 0002, South Africa; [email protected] (M.A.v.d.N.); [email protected] (E.T.S.); [email protected] (A.H.); brenda.wingfi[email protected] (B.D.W.) 2 Department of Plant Science, University of Manitoba, 222 Agriculture Building, Winnipeg, MB R3T 2N2, Canada 3 Biotechnology Platform, Agricultural Research Council (ARC), Onderstepoort Campus, Pretoria 0110, South Africa 4 Department of Mycology and Microbiology, University of Tartu, 14A Ravila, 50411 Tartu, Estonia; [email protected] * Correspondence: [email protected]; Fax: +1-204-474-7528 Abstract: Terpenes represent the biggest group of natural compounds on earth. This large class of organic hydrocarbons is distributed among all cellular organisms, including fungi. The different classes of terpenes produced by fungi are mono, sesqui, di- and triterpenes, although triterpene ergosterol is the main sterol identified in cell membranes of these organisms. The availability of genomic data from members in the Ceratocystidaceae enabled the detection and characterization of the genes encoding the enzymes in the mevalonate and ergosterol biosynthetic pathways. Using Citation: Sayari, M.; van der Nest, a bioinformatics approach, fungal orthologs of sterol biosynthesis genes in nine different species M.A.; Steenkamp, E.T.; Rahimlou, S.; of the Ceratocystidaceae were identified. -

The Impact of Transcription on Metabolism in Prostate and Breast Cancers
25 9 Endocrine-Related N Poulose et al. From hormones to fats and 25:9 R435–R452 Cancer back REVIEW The impact of transcription on metabolism in prostate and breast cancers Ninu Poulose1, Ian G Mills1,2,* and Rebecca E Steele1,* 1Centre for Cancer Research and Cell Biology, Queen’s University of Belfast, Belfast, UK 2Nuffield Department of Surgical Sciences, John Radcliffe Hospital, University of Oxford, Oxford, UK Correspondence should be addressed to I G Mills: [email protected] *(I G Mills and R E Steele contributed equally to this work) Abstract Metabolic dysregulation is regarded as an important driver in cancer development and Key Words progression. The impact of transcriptional changes on metabolism has been intensively f androgen studied in hormone-dependent cancers, and in particular, in prostate and breast cancer. f androgen receptor These cancers have strong similarities in the function of important transcriptional f breast drivers, such as the oestrogen and androgen receptors, at the level of dietary risk and f oestrogen epidemiology, genetics and therapeutically. In this review, we will focus on the function f endocrine therapy of these nuclear hormone receptors and their downstream impact on metabolism, with a resistance particular focus on lipid metabolism. We go on to discuss how lipid metabolism remains dysregulated as the cancers progress. We conclude by discussing the opportunities that this presents for drug repurposing, imaging and the development and testing of new Endocrine-Related Cancer therapeutics and treatment combinations. (2018) 25, R435–R452 Introduction: prostate and breast cancer Sex hormones act through nuclear hormone receptors Early-stage PCa is dependent on androgens for survival and induce distinct transcriptional programmes essential and can be treated by androgen deprivation therapy; to male and female physiology. -

33 34 35 Lipid Synthesis Laptop
BI/CH 422/622 Liver cytosol ANABOLISM OUTLINE: Photosynthesis Carbohydrate Biosynthesis in Animals Biosynthesis of Fatty Acids and Lipids Fatty Acids Triacylglycerides contrasts Membrane lipids location & transport Glycerophospholipids Synthesis Sphingolipids acetyl-CoA carboxylase Isoprene lipids: fatty acid synthase Ketone Bodies ACP priming 4 steps Cholesterol Control of fatty acid metabolism isoprene synth. ACC Joining Reciprocal control of b-ox Cholesterol Synth. Diversification of fatty acids Fates Eicosanoids Cholesterol esters Bile acids Prostaglandins,Thromboxanes, Steroid Hormones and Leukotrienes Metabolism & transport Control ANABOLISM II: Biosynthesis of Fatty Acids & Lipids Lipid Fat Biosynthesis Catabolism Fatty Acid Fatty Acid Synthesis Degradation Ketone body Utilization Isoprene Biosynthesis 1 Cholesterol and Steroid Biosynthesis mevalonate kinase Mevalonate to Activated Isoprenes • Two phosphates are transferred stepwise from ATP to mevalonate. • A third phosphate from ATP is added at the hydroxyl, followed by decarboxylation and elimination catalyzed by pyrophospho- mevalonate decarboxylase creates a pyrophosphorylated 5-C product: D3-isopentyl pyrophosphate (IPP) (isoprene). • Isomerization to a second isoprene dimethylallylpyrophosphate (DMAPP) gives two activated isoprene IPP compounds that act as precursors for D3-isopentyl pyrophosphate Isopentyl-D-pyrophosphate all of the other lipids in this class isomerase DMAPP Cholesterol and Steroid Biosynthesis mevalonate kinase Mevalonate to Activated Isoprenes • Two phosphates -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -
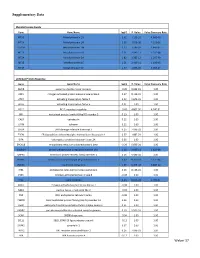
Weber 37 Supplementary Data
Supplementary Data Metallothionein Family Gene Gene Name logFC P. Value False Discovery Rate MT1G Metallothionein 1G 2.62 1.19E-10 4.66E-08 MT1H Metallothionein 1H 2.58 4.87E-08 7.12E-06 MT1M Metallothionein 1M 2.23 1.18E-07 1.44E-05 MT1X Metallothionein 1X 2.21 2.64E-11 1.25E-08 MT2A Metallothionein 2A 1.82 1.38E-12 1.26E-09 MT1E Metallothionein 1E 1.51 2.41E-11 1.18E-08 MT1F Metallothionein 1F 1.47 1.85E-09 4.09E-07 Unfolded Protein Response Gene Gene Name logFC P. Value False Discovery Rate AMFR autocrine motility factor receptor -0.20 9.94E-01 1.00 ASK1 mitogen-activated protein kinase kinase kinase 5 0.27 8.11E-01 1.00 ATF4 activating transcription factor 4 0.22 9.67E-01 1.00 ATF6 activating transcription factor 6 0.01 1.00 1.00 BCL2 BCL2, apoptosis regulator -0.43 4.80E-02 4.24E-01 BIP heat shock protein family A (Hsp70) member 5 0.13 1.00 1.00 CALR calreticulin 0.12 1.00 1.00 CANX calnexin 0.12 1.00 1.00 CHOP DNA damage inducible transcript 3 0.25 7.99E-01 1.00 EDEM ER degradation enhancing alpha-mannosidase like protein 1 0.37 1.88E-01 1.00 EIFA eukaryotic translation initiation factor 2A 0.06 1.00 1.00 ERO1LB endoplasmic reticulum oxidoreductase 1 beta -0.29 5.69E-01 1.00 GADD34 protein phosphatase 1 regulatory subunit 15A 1.18 3.66E-11 1.62E-08 GRP94 heat shock protein 90 beta family member 1 0.16 9.98E-01 1.00 HSPH1 heat shock protein family H (Hsp110) member 1 0.62 9.70E-06 5.12E-04 INSIG1 insulin induced gene 1 1.69 8.47E-18 3.88E-14 IRE1 endoplasmic reticulum to nucleus signaling 1 0.26 8.13E-01 1.00 JNK1 mitogen-activated -

Supplement 1A Steffensen Et
Liver Wild-type Knockout C T C T 1 2 4 7 8 9 1 2 4 5 7 9 1 1 1 1 1 1 C C C T T T C C C T T T W W W W W W K K K K K K IMAGE:793166 RIKEN cDNA 6720463E02 gene IMAGE:1447421 ESTs, Weakly similar to ZF37 MOUSE ZINC FINGER PROTEIN 37 [M.musculus] IMAGE:934291 RIKEN cDNA 2810418N01 gene IMAGE:1247525 small EDRK-rich f2actor IMAGE:1449402 expressed sequence AW321064 IMAGE:1279847 ESTs IMAGE:518737 expressed sequence AW049941 IMAGE:860231 a disintegrin and metalloproteinase domain 17 IMAGE:642836 CD86 antigen IMAGE:1003885 phosphoribosyl pyrophosphate sy1nthetase IMAGE:524862 RIKEN cDNA 5730469D23 gene IMAGE:1264473 protein inhibitor of activat1ed STAT IMAGE:847035 RIKEN cDNA 4833422F06 gene IMAGE:374550 requiem IMAGE:976520 nuclear receptor coact4ivator IMAGE:1264311 Unknown IMAGE:976735 expressed sequence AI987692 IMAGE:976659 cathepsLin IMAGE:1477580 RIKEN cDNA 1600010J02 gene IMAGE:1277168 ribosomal protein, large, P1 IMAGE:524842 RIKEN cDNA 0710008D09 gene IMAGE:373019 split hand/foot delete1d gene IMAGE:404428 expressed sequence AI413851 IMAGE:619810 RIKEN cDNA 1700003F10 gene IMAGE:1749558 caspase 3, apoptosis related cysteine protease IMAGE:718718 RIKEN cDNA 2810003F23 gene IMAGE:819789 Unknown IMAGE:524474 ATP-binding cassette, sub-family A ABC1, member IMAGE:804950 Mus musculus, Similar to ribosomal protein S20, clone MGC:6876 IMAGE:2651405, mRNA, complete cds IMAGE:806143 gap junction membrane channel prot2ein beta IMAGE:1745887 expressed sequence AI836376 IMAGE:779426 RIKEN cDNA 5230400G24 gene IMAGE:1125615 Unknown IMAGE:535025 DNA -

Appendix 2. Significantly Differentially Regulated Genes in Term Compared with Second Trimester Amniotic Fluid Supernatant
Appendix 2. Significantly Differentially Regulated Genes in Term Compared With Second Trimester Amniotic Fluid Supernatant Fold Change in term vs second trimester Amniotic Affymetrix Duplicate Fluid Probe ID probes Symbol Entrez Gene Name 1019.9 217059_at D MUC7 mucin 7, secreted 424.5 211735_x_at D SFTPC surfactant protein C 416.2 206835_at STATH statherin 363.4 214387_x_at D SFTPC surfactant protein C 295.5 205982_x_at D SFTPC surfactant protein C 288.7 1553454_at RPTN repetin solute carrier family 34 (sodium 251.3 204124_at SLC34A2 phosphate), member 2 238.9 206786_at HTN3 histatin 3 161.5 220191_at GKN1 gastrokine 1 152.7 223678_s_at D SFTPA2 surfactant protein A2 130.9 207430_s_at D MSMB microseminoprotein, beta- 99.0 214199_at SFTPD surfactant protein D major histocompatibility complex, class II, 96.5 210982_s_at D HLA-DRA DR alpha 96.5 221133_s_at D CLDN18 claudin 18 94.4 238222_at GKN2 gastrokine 2 93.7 1557961_s_at D LOC100127983 uncharacterized LOC100127983 93.1 229584_at LRRK2 leucine-rich repeat kinase 2 HOXD cluster antisense RNA 1 (non- 88.6 242042_s_at D HOXD-AS1 protein coding) 86.0 205569_at LAMP3 lysosomal-associated membrane protein 3 85.4 232698_at BPIFB2 BPI fold containing family B, member 2 84.4 205979_at SCGB2A1 secretoglobin, family 2A, member 1 84.3 230469_at RTKN2 rhotekin 2 82.2 204130_at HSD11B2 hydroxysteroid (11-beta) dehydrogenase 2 81.9 222242_s_at KLK5 kallikrein-related peptidase 5 77.0 237281_at AKAP14 A kinase (PRKA) anchor protein 14 76.7 1553602_at MUCL1 mucin-like 1 76.3 216359_at D MUC7 mucin 7, -

Aquatic Toxicology 176 (2016) 116–127
Aquatic Toxicology 176 (2016) 116–127 Contents lists available at ScienceDirect Aquatic Toxicology j ournal homepage: www.elsevier.com/locate/aquatox Linking the response of endocrine regulated genes to adverse effects on sex differentiation improves comprehension of aromatase inhibition in a Fish Sexual Development Test a,∗ b b b Elke Muth-Köhne , Kathi Westphal-Settele , Jasmin Brückner , Sabine Konradi , c a a,1 d,1 Viktoria Schiller , Christoph Schäfers , Matthias Teigeler , Martina Fenske a Fraunhofer IME, Department of Ecotoxicology, Auf dem Aberg 1, 57392 Schmallenberg, Germany b German Environment Agency (UBA), Woerlitzer Platz 1, 06844 Dessau, Germany c Fraunhofer IME, Attract Group UNIFISH, Forckenbeckstraße 6, 52074 Aachen, Germany d Fraunhofer IME, Project Group Translational Medicine and Pharmacology TMP, Forckenbeckstraße 6, 52074 Aachen, Germany a r a t i c l e i n f o b s t r a c t Article history: The Fish Sexual Development Test (FSDT) is a non-reproductive test to assess adverse effects of endocrine Received 23 December 2015 disrupting chemicals. With the present study it was intended to evaluate whether gene expression end- Received in revised form 13 April 2016 points would serve as predictive markers of endocrine disruption in a FSDT. For proof-of-concept, a FSDT Accepted 19 April 2016 according to the OECD TG 234 was conducted with the non-steroidal aromatase inhibitor fadrozole (test Available online 20 April 2016 concentrations: 10 g/L, 32 g/L, 100 g/L) using zebrafish (Danio rerio). Gene expression analyses using quantitative RT-PCR were included at 48 h, 96 h, 28 days and 63 days post fertilization (hpf, dpf). -

Update on Pathogenesis of Primary
177:3 C E Flück Update primary 177:3 R99–R111 Review adrenal insufficiency MECHANISMS IN ENDOCRINOLOGY Update on pathogenesis of primary adrenal insufficiency: beyond steroid enzyme deficiency and autoimmune adrenal destruction Correspondence Christa E Flück should be addressed Departments of Pediatrics and Clinical Research, Bern University Children’s Hospital Inselspital, University of Bern, to C E Flück Bern, Switzerland Email [email protected] Abstract Primary adrenal insufficiency (PAI) is potentially life threatening, but rare. In children, genetic defects prevail whereas adults suffer more often from acquired forms of PAI. The spectrum of genetic defects has increased in recent years with the use of next-generation sequencing methods and now has reached far beyond genetic defects in all known enzymes of adrenal steroidogenesis. Cofactor disorders such as P450 oxidoreductase (POR) deficiency manifesting as a complex form of congenital adrenal hyperplasia with a broad clinical phenotype have come to the fore. In patients with isolated familial glucocorticoid deficiency (FGD), in which no mutations in the genes for the ACTH receptor (MC2R) or its accessory protein MRAP have been found, non-classic steroidogenic acute regulatory protein (StAR) and CYP11A1 mutations have been described; and more recently novel mutations in genes such as nicotinamide nucleotide transhydrogenase (NNT) and thioredoxin reductase 2 (TRXR2) involved in the maintenance of the mitochondrial redox potential and generation of NADPH important for steroidogenesis and ROS detoxication have been discovered. European Journal European of Endocrinology In addition, whole exome sequencing approach also solved the genetics of some syndromic forms of PAI including IMAGe syndrome (CDKN1C), Irish traveler syndrome (MCM4), MIRAGE syndrome (SAMD9); and most recently a syndrome combining FGD with steroid-resistant nephrotic syndrome and ichthyosis caused by mutations in the gene for sphingosine-1-phosphate lyase 1 (SGPL1). -

A Key Mammalian Cholesterol Synthesis Enzyme, Squalene Monooxygenase, Is Allosterically Stabilized by Its Substrate
A key mammalian cholesterol synthesis enzyme, squalene monooxygenase, is allosterically stabilized by its substrate Hiromasa Yoshiokaa, Hudson W. Coatesb, Ngee Kiat Chuab, Yuichi Hashimotoa, Andrew J. Brownb,1, and Kenji Ohganea,c,1 aInstitute for Quantitative Biosciences, The University of Tokyo, 113-0032 Tokyo, Japan; bSchool of Biotechnology and Biomolecular Sciences, University of New South Wales, Sydney, NSW 2052, Australia; and cSynthetic Organic Chemistry Laboratory, RIKEN, 351-0198 Saitama, Japan Edited by Brenda A. Schulman, Max Planck Institute of Biochemistry, Martinsried, Germany, and approved February 18, 2020 (received for review September 20, 2019) Cholesterol biosynthesis is a high-cost process and, therefore, is the cognate E3 ligase for SM (9, 10), catalyzing its ubiquitination tightly regulated by both transcriptional and posttranslational in the N100 region (11). negative feedback mechanisms in response to the level of cellular Despite its importance in cholesterol-accelerated degradation, cholesterol. Squalene monooxygenase (SM, also known as squalene the structure and function of the SM-N100 region has not been epoxidase or SQLE) is a rate-limiting enzyme in the cholesterol fully resolved. Thus far, biochemical analyses have revealed the biosynthetic pathway and catalyzes epoxidation of squalene. The presence of a reentrant loop anchoring SM to the ER membrane stability of SM is negatively regulated by cholesterol via its and an amphipathic helix required for cholesterol-accelerated N-terminal regulatory domain (SM-N100). In this study, using a degradation (7, 8), while X-ray crystallography has revealed the SM-luciferase fusion reporter cell line, we performed a chemical architecture of the catalytic domain of SM (12). However, the genetics screen that identified inhibitors of SM itself as up-regulators highly hydrophobic and disordered nature of the N100 region of SM. -

Steroidal Triterpenes of Cholesterol Synthesis
Molecules 2013, 18, 4002-4017; doi:10.3390/molecules18044002 OPEN ACCESS molecules ISSN 1420-3049 www.mdpi.com/journal/molecules Review Steroidal Triterpenes of Cholesterol Synthesis Jure Ačimovič and Damjana Rozman * Centre for Functional Genomics and Bio-Chips, Faculty of Medicine, Institute of Biochemistry, University of Ljubljana, Zaloška 4, Ljubljana SI-1000, Slovenia; E-Mail: [email protected] * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +386-1-543-7591; Fax: +386-1-543-7588. Received: 18 February 2013; in revised form: 19 March 2013 / Accepted: 27 March 2013 / Published: 4 April 2013 Abstract: Cholesterol synthesis is a ubiquitous and housekeeping metabolic pathway that leads to cholesterol, an essential structural component of mammalian cell membranes, required for proper membrane permeability and fluidity. The last part of the pathway involves steroidal triterpenes with cholestane ring structures. It starts by conversion of acyclic squalene into lanosterol, the first sterol intermediate of the pathway, followed by production of 20 structurally very similar steroidal triterpene molecules in over 11 complex enzyme reactions. Due to the structural similarities of sterol intermediates and the broad substrate specificity of the enzymes involved (especially sterol-Δ24-reductase; DHCR24) the exact sequence of the reactions between lanosterol and cholesterol remains undefined. This article reviews all hitherto known structures of post-squalene steroidal triterpenes of cholesterol synthesis, their biological roles and the enzymes responsible for their synthesis. Furthermore, it summarises kinetic parameters of enzymes (Vmax and Km) and sterol intermediate concentrations from various tissues. Due to the complexity of the post-squalene cholesterol synthesis pathway, future studies will require a comprehensive meta-analysis of the pathway to elucidate the exact reaction sequence in different tissues, physiological or disease conditions. -

Integrating Data Mining, Network Pharmacology, and Molecular
Integrating Data Mining, Network Pharmacology, and Molecular Docking Verication to Investigate the Molecular Mechanism of Traditional Chinese Medicine Prescriptions for Treating Male Infertility Xue Bai Beijing University of Chinese Medicine Yibo Tang Beijing University of Chinese Medicine Qiang Li Beijing University of Chinese Medicine Guimin Liu Beijing University of Chinese Medicine Dan Liu Beijing University of Chinese Medicine Xiaolei Fan Beijing University of Chinese Medicine Zhejun Liu Beijing University of Chinese Medicine Shujun Yu Beijing University of Chinese Medicine Tian Tang Beijing University of Chinese Medicine Shuyan Wang Beijing University of Chinese Medicine Lingru Li Beijing University of Chinese Medicine Kailin Zhou Beijing University of Chinese Medicine Yanfei Zheng Beijing University of Chinese Medicine Zhenquan Liu ( [email protected] ) Research Page 1/42 Keywords: Traditional Chinese medicine, male infertility, data mining, network pharmacology, molecular docking Posted Date: March 4th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-264555/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 2/42 Abstract Background: Male infertility (MI) affects almost 5% adult men worldwide, and 75% of these cases are unexplained idiopathic. There are limitations in the current treatment due to the unclear mechanism of MI, which highlight the urgent need for a more effective strategy or drug. Traditional Chinese Medicine (TCM) prescriptions have been used to treat MI for thousands of years, but their molecular mechanism is not well dened. Methods: Aiming at revealing the molecular mechanism of TCM prescriptions on MI, a comprehensive strategy integrating data mining, network pharmacology, and molecular docking verication was performed.