SUPPLEMENTARY MATERIAL Supplementary Table 1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Next-Generation Sequencing of Representational Difference Analysis
Planta DOI 10.1007/s00425-017-2657-0 ORIGINAL ARTICLE Next-generation sequencing of representational difference analysis products for identification of genes involved in diosgenin biosynthesis in fenugreek (Trigonella foenum-graecum) 1 1 1 1 Joanna Ciura • Magdalena Szeliga • Michalina Grzesik • Mirosław Tyrka Received: 19 August 2016 / Accepted: 30 January 2017 Ó The Author(s) 2017. This article is published with open access at Springerlink.com Abstract Within the transcripts related to sterol and steroidal Main conclusion Representational difference analysis saponin biosynthesis, we discovered novel candidate of cDNA was performed and differential products were genes of diosgenin biosynthesis and validated their sequenced and annotated. Candidate genes involved in expression using quantitative RT-PCR analysis. Based on biosynthesis of diosgenin in fenugreek were identified. these findings, we supported the idea that diosgenin is Detailed mechanism of diosgenin synthesis was biosynthesized from cycloartenol via cholesterol. This is proposed. the first report on the next-generation sequencing of cDNA-RDA products. Analysis of the transcriptomes Fenugreek (Trigonella foenum-graecum L.) is a valuable enriched in low copy sequences contributed substantially medicinal and crop plant. It belongs to Fabaceae family to our understanding of the biochemical pathways of and has a unique potential to synthesize valuable steroidal steroid synthesis in fenugreek. saponins, e.g., diosgenin. Elicitation (methyl jasmonate) and precursor feeding (cholesterol and squalene) were Keywords Diosgenin Á Next-generation sequencing Á used to enhance the content of sterols and steroidal Phytosterols Á Representational difference analysis of sapogenins in in vitro grown plants for representational cDNA Á Steroidal saponins Á Transcriptome user-friendly difference analysis of cDNA (cDNA-RDA). -

Characterization of the Ergosterol Biosynthesis Pathway in Ceratocystidaceae
Journal of Fungi Article Characterization of the Ergosterol Biosynthesis Pathway in Ceratocystidaceae Mohammad Sayari 1,2,*, Magrieta A. van der Nest 1,3, Emma T. Steenkamp 1, Saleh Rahimlou 4 , Almuth Hammerbacher 1 and Brenda D. Wingfield 1 1 Department of Biochemistry, Genetics and Microbiology, Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, Pretoria 0002, South Africa; [email protected] (M.A.v.d.N.); [email protected] (E.T.S.); [email protected] (A.H.); brenda.wingfi[email protected] (B.D.W.) 2 Department of Plant Science, University of Manitoba, 222 Agriculture Building, Winnipeg, MB R3T 2N2, Canada 3 Biotechnology Platform, Agricultural Research Council (ARC), Onderstepoort Campus, Pretoria 0110, South Africa 4 Department of Mycology and Microbiology, University of Tartu, 14A Ravila, 50411 Tartu, Estonia; [email protected] * Correspondence: [email protected]; Fax: +1-204-474-7528 Abstract: Terpenes represent the biggest group of natural compounds on earth. This large class of organic hydrocarbons is distributed among all cellular organisms, including fungi. The different classes of terpenes produced by fungi are mono, sesqui, di- and triterpenes, although triterpene ergosterol is the main sterol identified in cell membranes of these organisms. The availability of genomic data from members in the Ceratocystidaceae enabled the detection and characterization of the genes encoding the enzymes in the mevalonate and ergosterol biosynthetic pathways. Using Citation: Sayari, M.; van der Nest, a bioinformatics approach, fungal orthologs of sterol biosynthesis genes in nine different species M.A.; Steenkamp, E.T.; Rahimlou, S.; of the Ceratocystidaceae were identified. -

Relating Metatranscriptomic Profiles to the Micropollutant
1 Relating Metatranscriptomic Profiles to the 2 Micropollutant Biotransformation Potential of 3 Complex Microbial Communities 4 5 Supporting Information 6 7 Stefan Achermann,1,2 Cresten B. Mansfeldt,1 Marcel Müller,1,3 David R. Johnson,1 Kathrin 8 Fenner*,1,2,4 9 1Eawag, Swiss Federal Institute of Aquatic Science and Technology, 8600 Dübendorf, 10 Switzerland. 2Institute of Biogeochemistry and Pollutant Dynamics, ETH Zürich, 8092 11 Zürich, Switzerland. 3Institute of Atmospheric and Climate Science, ETH Zürich, 8092 12 Zürich, Switzerland. 4Department of Chemistry, University of Zürich, 8057 Zürich, 13 Switzerland. 14 *Corresponding author (email: [email protected] ) 15 S.A and C.B.M contributed equally to this work. 16 17 18 19 20 21 This supporting information (SI) is organized in 4 sections (S1-S4) with a total of 10 pages and 22 comprises 7 figures (Figure S1-S7) and 4 tables (Table S1-S4). 23 24 25 S1 26 S1 Data normalization 27 28 29 30 Figure S1. Relative fractions of gene transcripts originating from eukaryotes and bacteria. 31 32 33 Table S1. Relative standard deviation (RSD) for commonly used reference genes across all 34 samples (n=12). EC number mean fraction bacteria (%) RSD (%) RSD bacteria (%) RSD eukaryotes (%) 2.7.7.6 (RNAP) 80 16 6 nda 5.99.1.2 (DNA topoisomerase) 90 11 9 nda 5.99.1.3 (DNA gyrase) 92 16 10 nda 1.2.1.12 (GAPDH) 37 39 6 32 35 and indicates not determined. 36 37 38 39 S2 40 S2 Nitrile hydration 41 42 43 44 Figure S2: Pearson correlation coefficients r for rate constants of bromoxynil and acetamiprid with 45 gene transcripts of ECs describing nucleophilic reactions of water with nitriles. -

( 12 ) United States Patent
US010208322B2 (12 ) United States Patent ( 10 ) Patent No. : US 10 ,208 , 322 B2 Coelho et al. (45 ) Date of Patent: * Feb . 19, 2019 ( 54 ) IN VIVO AND IN VITRO OLEFIN ( 56 ) References Cited CYCLOPROPANATION CATALYZED BY HEME ENZYMES U . S . PATENT DOCUMENTS 3 , 965 ,204 A 6 / 1976 Lukas et al. (71 ) Applicant: California Institute of Technology , 4 , 243 ,819 A 1 / 1981 Henrick Pasadena , CA (US ) 5 ,296 , 595 A 3 / 1994 Doyle 5 , 703 , 246 A 12 / 1997 Aggarwal et al. 7 , 226 , 768 B2 6 / 2007 Farinas et al. ( 72 ) Inventors : Pedro S . Coelho , Los Angeles, CA 7 , 267 , 949 B2 9 / 2007 Richards et al . (US ) ; Eric M . Brustad , Durham , NC 7 ,625 ,642 B2 12 / 2009 Matsutani et al. (US ) ; Frances H . Arnold , La Canada , 7 ,662 , 969 B2 2 / 2010 Doyle et al. CA (US ) ; Zhan Wang , San Jose , CA 7 ,863 ,030 B2 1 / 2011 Arnold (US ) ; Jared C . Lewis , Chicago , IL 8 ,247 ,430 B2 8 / 2012 Yuan 8 , 993 , 262 B2 * 3 / 2015 Coelho . .. .. .. • * • C12P 7 /62 (US ) 435 / 119 9 ,399 , 762 B26 / 2016 Farwell et al . (73 ) Assignee : California Institute of Technology , 9 , 493 ,799 B2 * 11 /2016 Coelho .. C12P 7162 Pasadena , CA (US ) 2006 / 0030718 AL 2 / 2006 Zhang et al. 2006 / 0111347 A1 5 / 2006 Askew , Jr . et al. 2007 /0276013 AL 11 /2007 Ebbinghaus et al . ( * ) Notice : Subject to any disclaimer , the term of this 2009 /0238790 A2 9 /2009 Sommadosi et al. patent is extended or adjusted under 35 2010 / 0056806 A1 3 / 2010 Warren U . -

Aquatic Toxicology 176 (2016) 116–127
Aquatic Toxicology 176 (2016) 116–127 Contents lists available at ScienceDirect Aquatic Toxicology j ournal homepage: www.elsevier.com/locate/aquatox Linking the response of endocrine regulated genes to adverse effects on sex differentiation improves comprehension of aromatase inhibition in a Fish Sexual Development Test a,∗ b b b Elke Muth-Köhne , Kathi Westphal-Settele , Jasmin Brückner , Sabine Konradi , c a a,1 d,1 Viktoria Schiller , Christoph Schäfers , Matthias Teigeler , Martina Fenske a Fraunhofer IME, Department of Ecotoxicology, Auf dem Aberg 1, 57392 Schmallenberg, Germany b German Environment Agency (UBA), Woerlitzer Platz 1, 06844 Dessau, Germany c Fraunhofer IME, Attract Group UNIFISH, Forckenbeckstraße 6, 52074 Aachen, Germany d Fraunhofer IME, Project Group Translational Medicine and Pharmacology TMP, Forckenbeckstraße 6, 52074 Aachen, Germany a r a t i c l e i n f o b s t r a c t Article history: The Fish Sexual Development Test (FSDT) is a non-reproductive test to assess adverse effects of endocrine Received 23 December 2015 disrupting chemicals. With the present study it was intended to evaluate whether gene expression end- Received in revised form 13 April 2016 points would serve as predictive markers of endocrine disruption in a FSDT. For proof-of-concept, a FSDT Accepted 19 April 2016 according to the OECD TG 234 was conducted with the non-steroidal aromatase inhibitor fadrozole (test Available online 20 April 2016 concentrations: 10 g/L, 32 g/L, 100 g/L) using zebrafish (Danio rerio). Gene expression analyses using quantitative RT-PCR were included at 48 h, 96 h, 28 days and 63 days post fertilization (hpf, dpf). -

Geneid T-Test Q-Val Gene Symbol Gene Title GO Biological Process
Supplementary Table 2. Genes selected by FADA as the most discriminant between tumoral and normal prostate samples GO Biological Process GO Molecular Function GO Cellular Component GeneID T-test q-val Gene Symbol Gene Title Description Description Description Pathway serine-type endopeptidase inhibitor activity /// endopeptidase inhibitor serpin peptidase inhibitor, clade activity /// serine-type endopeptidase 213572_s_at -6.753 2.213E-05 SERPINB1 B (ovalbumin), member 1 --- inhibitor activity cytoplasm --- ion transport /// cellular defense response /// positive regulation of T-cell, immune regulator 1, cell proliferation /// proton plasma membrane /// integral to ATPase, H+ transporting, transport /// transport /// proton transporter activity /// hydrogen ion plasma membrane /// membrane /// 204158_s_at -5.729 8.585E-05 TCIRG1 lysosomal V0 subunit A3 transport transporter activity integral to membrane --- chemotaxis /// cell adhesion /// homophilic cell adhesion /// nervous system development /// cell differentiation /// positive roundabout, axon guidance regulation of axonogenesis /// receptor activity /// axon guidance integral to plasma membrane /// cell receptor, homolog 1 development /// nervous system receptor activity /// identical protein surface /// membrane /// integral to 213194_at -6.168 4.441E-05 ROBO1 (Drosophila) development binding /// protein binding membrane --- structural molecule activity /// nucleus /// intermediate filament /// protein binding /// microtubule cytoplasmic dynein complex /// 203411_s_at -5.297 1.636E-04 -

Steroidal Triterpenes of Cholesterol Synthesis
Molecules 2013, 18, 4002-4017; doi:10.3390/molecules18044002 OPEN ACCESS molecules ISSN 1420-3049 www.mdpi.com/journal/molecules Review Steroidal Triterpenes of Cholesterol Synthesis Jure Ačimovič and Damjana Rozman * Centre for Functional Genomics and Bio-Chips, Faculty of Medicine, Institute of Biochemistry, University of Ljubljana, Zaloška 4, Ljubljana SI-1000, Slovenia; E-Mail: [email protected] * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +386-1-543-7591; Fax: +386-1-543-7588. Received: 18 February 2013; in revised form: 19 March 2013 / Accepted: 27 March 2013 / Published: 4 April 2013 Abstract: Cholesterol synthesis is a ubiquitous and housekeeping metabolic pathway that leads to cholesterol, an essential structural component of mammalian cell membranes, required for proper membrane permeability and fluidity. The last part of the pathway involves steroidal triterpenes with cholestane ring structures. It starts by conversion of acyclic squalene into lanosterol, the first sterol intermediate of the pathway, followed by production of 20 structurally very similar steroidal triterpene molecules in over 11 complex enzyme reactions. Due to the structural similarities of sterol intermediates and the broad substrate specificity of the enzymes involved (especially sterol-Δ24-reductase; DHCR24) the exact sequence of the reactions between lanosterol and cholesterol remains undefined. This article reviews all hitherto known structures of post-squalene steroidal triterpenes of cholesterol synthesis, their biological roles and the enzymes responsible for their synthesis. Furthermore, it summarises kinetic parameters of enzymes (Vmax and Km) and sterol intermediate concentrations from various tissues. Due to the complexity of the post-squalene cholesterol synthesis pathway, future studies will require a comprehensive meta-analysis of the pathway to elucidate the exact reaction sequence in different tissues, physiological or disease conditions. -

Expression of Selected Genes Involved in Steroidogenesis in the Course of Enucleation-Induced Rat Adrenal Regeneration
INTERNATIONAL JOURNAL OF MOLECULAR MEDICINE 33: 613-623, 2014 Expression of selected genes involved in steroidogenesis in the course of enucleation-induced rat adrenal regeneration MARIANNA TYCZEWSKA, MARCIN RUCINSKI, AGNIESZKA ZIOLKOWSKA, MARCIN TREJTER, MARTA SZYSZKA and LUDWIK K. MALENDOWICZ Department of Histology and Embryology, Poznan University of Medical Sciences, Poznan, Poland Received October 28, 2013; Accepted December 6, 2013 DOI: 10.3892/ijmm.2013.1599 Abstract. The enucleation-induced (EI) rapid proliferation however, throughout the entire experimental period, there were of adrenocortical cells is followed by their differentiation, no statistically significant differences observed. After the initial the degree of which may be characterized by the expression decrease in steroidogenic factor 1 (Sf-1) mRNA levels observed of genes directly and indirectly involved in steroid hormone on the 1st day of the experiment, a marked upregulation in its biosynthesis. In this study, out of 30,000 transcripts of genes expression was observed from there on. Data from the current identified by means of Affymetrix Rat Gene 1.1 ST Array, we study strongly suggest the role of Fabp6, Lipe and Soat1 in aimed to select genes (either up- or downregulated) involved supplying substrates of regenerating adrenocortical cells for in steroidogenesis in the course of enucleation-induced adrenal steroid synthesis. Our results indicate that during the first days regeneration. On day 1, we found 32 genes with altered of adrenal regeneration, intense synthesis of cholesterol may expression levels, 15 were upregulated and 17 were down- occur, which is then followed by its conversion into cholesteryl regulated [i.e., 3β-hydroxysteroid dehydrogenase (Hsd3β), esters. -
Figures Intermediary Ontogeny.Pptx
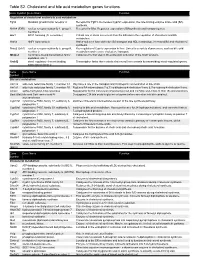
Table S2. Cholesterol and bile acid metabolism genes functions. Gene Symbol Gene Name Function Regulation of cholesterol and/or bile acid metabolism Fgfr4 fibroblast growth factor receptor 4 Receptor for Fgf15. Decreases Cyp7a1 expression, the rate-limiting enzyme in bile acid (BA) synthesis. Nr1h4 (FXR) nuclear receptor subfamily 1, group H, Receptor for BAs. Regulates expression of BAsynthesis and transport genes. member 4 Arv1 ARV1 homolog (S. cerevisiae) Critical role in sterol movement from the ER and in the regulation of cholesterol and BA metabolism. Hnf1a HNF1 homeobox A Hnf1a-null mice have defective BA transport and HDL metabolism, increased BA and cholesterol synthesis. Nr5a2 (Lrh1) nuclear receptor subfamily 5, group A, Key regulator of Cyp7a expression in liver. Linked to a variety of processes, such as bile acid member 2 metabolism and reverse cholesterol transport. Mbtps1 membrane-bound transcription factor Catalyzes the first step in the proteolytic activation of the Srebf proteins. peptidase, site 1 Srebf2 sterol regulatory element binding Transcription factor that controls cholesterol homeostasis by transcribing sterol-regulated genes. transcription factor 2 Gene Gene Name Function Symbol Bile acid metabolism Akr1c6 aldo-keto reductase family 1, member C1 May have a role in the transport and intrahepatic concentration of bile acids. Akr1d1 aldo-keto reductase family 1, member D1 Reduces BA intermediates 7-α,12-α-dihydroxy-4-cholesten-3-one & 7-α-hydroxy-4-cholesten-3-one. Amacr alpha-methylacyl-CoA racemase Responsible for the conversion of pristanoyl-CoA and C27-bile acyl-CoAs to their (S)-stereoisomers. Baat (Bat) bile acid CoA: amino acid N- Conjugates C24 bile acids to glycine or taurine before excretion into bile canaliculi. -

The Use of Mutants and Inhibitors to Study Sterol Biosynthesis in Plants
bioRxiv preprint doi: https://doi.org/10.1101/784272; this version posted September 26, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 Title page 2 Title: The use of mutants and inhibitors to study sterol 3 biosynthesis in plants 4 5 Authors: Kjell De Vriese1,2, Jacob Pollier1,2,3, Alain Goossens1,2, Tom Beeckman1,2, Steffen 6 Vanneste1,2,4,* 7 Affiliations: 8 1: Department of Plant Biotechnology and Bioinformatics, Ghent University, Technologiepark 71, 9052 Ghent, 9 Belgium 10 2: VIB Center for Plant Systems Biology, VIB, Technologiepark 71, 9052 Ghent, Belgium 11 3: VIB Metabolomics Core, Technologiepark 71, 9052 Ghent, Belgium 12 4: Lab of Plant Growth Analysis, Ghent University Global Campus, Songdomunhwa-Ro, 119, Yeonsu-gu, Incheon 13 21985, Republic of Korea 14 15 e-mails: 16 K.D.V: [email protected] 17 J.P: [email protected] 18 A.G. [email protected] 19 T.B. [email protected] 20 S.V. [email protected] 21 22 *Corresponding author 23 Tel: +32 9 33 13844 24 Date of submission: sept 26th 2019 25 Number of Figures:3 in colour 26 Word count: 6126 27 28 1 bioRxiv preprint doi: https://doi.org/10.1101/784272; this version posted September 26, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

Dalmanol Biosyntheses Require Coupling of Two Separate Polyketide Gene Clusters
Showcasing research progress from Professor Ren Xiang Tan’s As featured in: laboratories at Nanjing University & Nanjing University of Chinese Medicine, Nanjing, China. Dalmanol biosyntheses require coupling of two separate polyketide gene clusters The biosynthesis of immunosuppressants (dalmanol A and acetodalmanol A) is characterized by the coupling of two separate (naphthalene- and chromane-encoding) gene clusters under the catalysis of unspecifi c enzyme(s) that may distribute widely in the fungal kingdom. The fi nding highlights an access to the inter-cluster orchestration derived chemodiversity of microbial secondary metabolites that remain to be a reliable source of leading compounds triggering the development of See Ren Xiang Tan et al., new medicines, agrochemicals, and other tool molecules. Chem. Sci., 2019, 10, 73. rsc.li/chemical-science Registered charity number: 207890 Chemical Science View Article Online EDGE ARTICLE View Journal | View Issue Dalmanol biosyntheses require coupling of two separate polyketide gene clusters† Cite this: Chem. Sci.,2019,10,73 a a bc a a All publication charges for this article Zhen Zhen Zhou, Hong Jie Zhu, Li Ping Lin, Xuan Zhang, Hui Ming Ge, have been paid for by the Royal Society Rui Hua Jiaoa and Ren Xiang Tan *ab of Chemistry Polyketide–polyketide hybrids are unique natural products with promising bioactivity, but the hybridization processes remain poorly understood. Herein, we present that the biosynthetic pathways of two immunosuppressants, dalmanol A and acetodalmanol A, result from an unspecific monooxygenase triggered hybridization of two distinct polyketide (naphthalene and chromane) biosynthetic gene clusters. The orchestration of the functional dimorphism of the polyketide synthase (ChrA) ketoreductase (KR) domain (shortened as ChrA KR) with that of the KR partner (ChrB) in the bioassembly line increases the polyketide diversity and allows the fungal generation of plant chromanes (e.g., noreugenin) and phloroglucinols (e.g., 2,4,6-trihydroxyacetophenone). -

Oxygen-Mediated Regulation of Cholesterol Synthesis
OXYGEN-MEDIATED REGULATION OF CHOLESTEROL SYNTHESIS THROUGH ACCELERATED DEGRADATION OF HMG COA REDUCTASE APPROVED BY SUPERVISORY COMMITTEE Russell DeBose-Boyd, Ph.D. Richard Bruick, Ph.D. Michael Brown, M.D. Zhijian Chen, Ph.D. George DeMartino, Ph.D. To my family For their love and support OXYGEN-MEDIATED REGULATION OF CHOLESTEROL SYNTHESIS THROUGH ACCELERATED DEGRADATION OF HMG COA REDUCTASE by ANDREW TUAN DUC NGUYEN DISSERTATION Presented to the Faculty of the Graduate School of Biomedical Sciences The University of Texas Southwestern Medical Center at Dallas In Partial Fulfillment of the Requirements For the Degree of DOCTOR OF PHILOSOPHY The University of Texas Southwestern Medical Center at Dallas Dallas, Texas May, 2009 Copyright by Andrew Tuan Duc Nguyen, 2009 All Rights Reserved OXYGEN-MEDIATED REGULATION OF CHOLESTEROL SYNTHESIS THROUGH ACCELERATED DEGRADATION OF HMG COA REDUCTASE Andrew Tuan Duc Nguyen, Ph.D. The University of Texas Southwestern Medical Center at Dallas, 2009 Supervising Professor: Russell A. DeBose-Boyd, Ph.D. Endoplasmic reticulum-associated degradation of the enzyme 3-hydroxy-3- methylglutaryl CoA reductase represents one mechanism by which cholesterol synthesis is controlled in mammalian cells. The key reaction in this degradation is binding of reductase to Insig proteins in the endoplasmic reticulum, which is stimulated by the methylated cholesterol precursors lanosterol and 24,25-dihydrolanosterol. Conversion of these sterols to cholesterol requires the removal of three methyl groups, which consumes v nine molecules of O2. Here, we report that oxygen deprivation (hypoxia) slows the rate of demethylation of lanosterol and its reduced metabolite 24,25-dihydrolanosterol, causing both sterols to accumulate in cells.