Combinatorial Biosynthesis of Sapogenins and Saponins in Saccharomyces Cerevisiae Using a C-16 Α Hydroxylase from Bupleurum
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

7Th International Conference on Countercurrent Chromatography, Hangzhou, August 6-8, 2012 Program
010 7th international conference on countercurrent chromatography, Hangzhou, August 6-8, 2012 Program January, August 6, 2012 8:30 – 9:00 Registration 9:00 – 9:10 Opening CCC 2012 Chairman: Prof. Qizhen Du 9:10 – 9:20 Welcome speech from the director of Zhejiang Gongshang University Session 1 – CCC Keynotes Chirman: Prof. Guoan Luo pH-zone-refining countercurrent chromatography : USA 09:20-09:50 Ito, Y. Origin, mechanism, procedure and applications K-1 Sutherland, I.*; Hewitson. P.; Scalable technology for the extraction of UK 09:50-10:20 Janaway, L.; Wood, P; pharmaceuticals (STEP): Outcomes from a year Ignatova, S. collaborative researchprogramme K-2 10:20-11:00 Tea Break with Poster & Exhibition session 1 France 11:00-11:30 Berthod, A. Terminology for countercurrent chromatography K-3 API recovery from pharmaceutical waste streams by high performance countercurrent UK 11:30-12:00 Ignatova, S.*; Sutherland, I. chromatography and intermittent countercurrent K-4 extraction 12:00-13:30 Lunch break 7th international conference on countercurrent chromatography, Hangzhou, August 6-8, 2012 January, August 6, 2012 Session 2 – CCC Instrumentation I Chirman: Prof. Ian Sutherland Pro, S.; Burdick, T.; Pro, L.; Friedl, W.; Novak, N.; Qiu, A new generation of countercurrent separation USA 13:30-14:00 F.; McAlpine, J.B., J. Brent technology O-1 Friesen, J.B.; Pauli, G.F.* Berthod, A.*; Faure, K.; A small volume hydrostatic CCC column for France 14:00-14:20 Meucci, J.; Mekaoui, N. full and quick solvent selection O-2 Construction of a HSCCC apparatus with Du, Q.B.; Jiang, H.; Yin, J.; column capacity of 12 or 15 liters and its China Xu, Y.; Du, W.; Li, B.; Du, application as flash countercurrent 14:20-14:40 O-3 Q.* chromatography in quick preparation of (-)-epicatechin 14:40-15:30 Tea Break with Poster & Exhibition session 2 Session 3 – CCC Instrumentation II Chirman: Prof. -

Characterization of the Ergosterol Biosynthesis Pathway in Ceratocystidaceae
Journal of Fungi Article Characterization of the Ergosterol Biosynthesis Pathway in Ceratocystidaceae Mohammad Sayari 1,2,*, Magrieta A. van der Nest 1,3, Emma T. Steenkamp 1, Saleh Rahimlou 4 , Almuth Hammerbacher 1 and Brenda D. Wingfield 1 1 Department of Biochemistry, Genetics and Microbiology, Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, Pretoria 0002, South Africa; [email protected] (M.A.v.d.N.); [email protected] (E.T.S.); [email protected] (A.H.); brenda.wingfi[email protected] (B.D.W.) 2 Department of Plant Science, University of Manitoba, 222 Agriculture Building, Winnipeg, MB R3T 2N2, Canada 3 Biotechnology Platform, Agricultural Research Council (ARC), Onderstepoort Campus, Pretoria 0110, South Africa 4 Department of Mycology and Microbiology, University of Tartu, 14A Ravila, 50411 Tartu, Estonia; [email protected] * Correspondence: [email protected]; Fax: +1-204-474-7528 Abstract: Terpenes represent the biggest group of natural compounds on earth. This large class of organic hydrocarbons is distributed among all cellular organisms, including fungi. The different classes of terpenes produced by fungi are mono, sesqui, di- and triterpenes, although triterpene ergosterol is the main sterol identified in cell membranes of these organisms. The availability of genomic data from members in the Ceratocystidaceae enabled the detection and characterization of the genes encoding the enzymes in the mevalonate and ergosterol biosynthetic pathways. Using Citation: Sayari, M.; van der Nest, a bioinformatics approach, fungal orthologs of sterol biosynthesis genes in nine different species M.A.; Steenkamp, E.T.; Rahimlou, S.; of the Ceratocystidaceae were identified. -

Sterols and Triterpenes: Antiviral Potential Supported by In-Silico Analysis
plants Review Sterols and Triterpenes: Antiviral Potential Supported by In-Silico Analysis Nourhan Hisham Shady 1,†, Khayrya A. Youssif 2,†, Ahmed M. Sayed 3 , Lassaad Belbahri 4 , Tomasz Oszako 5 , Hossam M. Hassan 3,6 and Usama Ramadan Abdelmohsen 1,7,* 1 Department of Pharmacognosy, Faculty of Pharmacy, Deraya University, Universities Zone, P.O. Box 61111, New Minia City, Minia 61519, Egypt; [email protected] 2 Department of Pharmacognosy, Faculty of Pharmacy, Modern University for Technology and Information, Cairo 11865, Egypt; [email protected] 3 Department of Pharmacognosy, Faculty of Pharmacy, Nahda University, Beni-Suef 62513, Egypt; [email protected] (A.M.S.); [email protected] (H.M.H.) 4 Laboratory of Soil Biology, University of Neuchatel, 2000 Neuchatel, Switzerland; [email protected] 5 Departement of Forest Protection, Forest Research Institute, 05-090 S˛ekocinStary, Poland; [email protected] 6 Department of Pharmacognosy, Faculty of Pharmacy, Beni-Suef University, Beni-Suef 62514, Egypt 7 Department of Pharmacognosy, Faculty of Pharmacy, Minia University, Minia 61519, Egypt * Correspondence: [email protected]; Tel.: +2-86-2347759 † Equal contribution. Abstract: The acute respiratory syndrome caused by the novel coronavirus (SARS-CoV-2) caused severe panic all over the world. The coronavirus (COVID-19) outbreak has already brought massive human suffering and major economic disruption and unfortunately, there is no specific treatment for COVID-19 so far. Herbal medicines and purified natural products can provide a rich resource for novel antiviral drugs. Therefore, in this review, we focused on the sterols and triterpenes as potential candidates derived from natural sources with well-reported in vitro efficacy against numerous types of viruses. -

Studies on Betalain Phytochemistry by Means of Ion-Pair Countercurrent Chromatography
STUDIES ON BETALAIN PHYTOCHEMISTRY BY MEANS OF ION-PAIR COUNTERCURRENT CHROMATOGRAPHY Von der Fakultät für Lebenswissenschaften der Technischen Universität Carolo-Wilhelmina zu Braunschweig zur Erlangung des Grades einer Doktorin der Naturwissenschaften (Dr. rer. nat.) genehmigte D i s s e r t a t i o n von Thu Tran Thi Minh aus Vietnam 1. Referent: Prof. Dr. Peter Winterhalter 2. Referent: apl. Prof. Dr. Ulrich Engelhardt eingereicht am: 28.02.2018 mündliche Prüfung (Disputation) am: 28.05.2018 Druckjahr 2018 Vorveröffentlichungen der Dissertation Teilergebnisse aus dieser Arbeit wurden mit Genehmigung der Fakultät für Lebenswissenschaften, vertreten durch den Mentor der Arbeit, in folgenden Beiträgen vorab veröffentlicht: Tagungsbeiträge T. Tran, G. Jerz, T.E. Moussa-Ayoub, S.K.EI-Samahy, S. Rohn und P. Winterhalter: Metabolite screening and fractionation of betalains and flavonoids from Opuntia stricta var. dillenii by means of High Performance Countercurrent chromatography (HPCCC) and sequential off-line injection to ESI-MS/MS. (Poster) 44. Deutscher Lebensmittelchemikertag, Karlsruhe (2015). Thu Minh Thi Tran, Tamer E. Moussa-Ayoub, Salah K. El-Samahy, Sascha Rohn, Peter Winterhalter und Gerold Jerz: Metabolite profile of betalains and flavonoids from Opuntia stricta var. dilleni by HPCCC and offline ESI-MS/MS. (Poster) 9. Countercurrent Chromatography Conference, Chicago (2016). Thu Tran Thi Minh, Binh Nguyen, Peter Winterhalter und Gerold Jerz: Recovery of the betacyanin celosianin II and flavonoid glycosides from Atriplex hortensis var. rubra by HPCCC and off-line ESI-MS/MS monitoring. (Poster) 9. Countercurrent Chromatography Conference, Chicago (2016). ACKNOWLEDGEMENT This PhD would not be done without the supports of my mentor, my supervisor and my family. -

Pressurized Liquid Extraction Applied for the Recovery of Phenolic Compounds from Beetroot Waste
Biocatalysis and Agricultural Biotechnology 21 (2019) 101353 Contents lists available at ScienceDirect Biocatalysis and Agricultural Biotechnology journal homepage: http://www.elsevier.com/locate/bab Pressurized liquid extraction applied for the recovery of phenolic compounds from beetroot waste Heloísa Fabian Battistella Lasta a, Lucas Lentz a, Luiz Gustavo Gonçalves Rodrigues a, Natalia� Mezzomo a, Luciano Vitali b, Sandra Regina Salvador Ferreira a,* a Chemical and Food Engineering Department, Federal University of Santa Catarina, EQA/UFSC, C.P. 476, CEP 88.040-900, Florianopolis,� SC, Brazil b Department of Chemistry, Federal University of Santa Catarina, Florianopolis,� SC, Brazil ARTICLE INFO ABSTRACT Keywords: Beetroot (Beta vulgaris L.) residues, leaves and stems, normally disposed as animal feed or compost, were Beta vulgaris L. residue investigated for the recovery of valuable compounds by pressurized liquid extraction (PLE) at temperature of � PLE 40 C and pressures of 7.5, 10 and 12.5 MPa, and flowrate of 3 mL min-1. This abundant residue, combined with Antioxidant potential the lack of related studies of beetroot waste by applying the PLE methods to obtain bioactive extracts is the main Phenolic compounds novelty of this work. Then, the objective was to explore the PLE process to enhance the value of the residues, Waste valorization evaluating process yield, total phenolic content (TPC) and antioxidant activity (DPPH, ABTS and FRAP methods) of the recovered extracts. Chemical composition was analyzed using LC-ESI-MS/MS and GC-MS analysis. Anti oxidant potential results suggest the recovered extracts are comparable with synthetic antioxidant (BHT). Ferulic acid, vitexin and sinapaldehyde, were the most abundant phenolic compounds obtained from the extracts. -

Aquatic Toxicology 176 (2016) 116–127
Aquatic Toxicology 176 (2016) 116–127 Contents lists available at ScienceDirect Aquatic Toxicology j ournal homepage: www.elsevier.com/locate/aquatox Linking the response of endocrine regulated genes to adverse effects on sex differentiation improves comprehension of aromatase inhibition in a Fish Sexual Development Test a,∗ b b b Elke Muth-Köhne , Kathi Westphal-Settele , Jasmin Brückner , Sabine Konradi , c a a,1 d,1 Viktoria Schiller , Christoph Schäfers , Matthias Teigeler , Martina Fenske a Fraunhofer IME, Department of Ecotoxicology, Auf dem Aberg 1, 57392 Schmallenberg, Germany b German Environment Agency (UBA), Woerlitzer Platz 1, 06844 Dessau, Germany c Fraunhofer IME, Attract Group UNIFISH, Forckenbeckstraße 6, 52074 Aachen, Germany d Fraunhofer IME, Project Group Translational Medicine and Pharmacology TMP, Forckenbeckstraße 6, 52074 Aachen, Germany a r a t i c l e i n f o b s t r a c t Article history: The Fish Sexual Development Test (FSDT) is a non-reproductive test to assess adverse effects of endocrine Received 23 December 2015 disrupting chemicals. With the present study it was intended to evaluate whether gene expression end- Received in revised form 13 April 2016 points would serve as predictive markers of endocrine disruption in a FSDT. For proof-of-concept, a FSDT Accepted 19 April 2016 according to the OECD TG 234 was conducted with the non-steroidal aromatase inhibitor fadrozole (test Available online 20 April 2016 concentrations: 10 g/L, 32 g/L, 100 g/L) using zebrafish (Danio rerio). Gene expression analyses using quantitative RT-PCR were included at 48 h, 96 h, 28 days and 63 days post fertilization (hpf, dpf). -

Triterpenoid Saponins Garai S* CSIR-Indian Institute of Chemical Biology, 4 Raja SC Mullick Road, Jadavpur, Kolkata-700032, India
s Chemis ct try u d & Garai, Nat Prod Chem Res 2014, 2:6 o r R P e s l e a r a DOI: 10.4172/2329-6836.1000148 r u t c h a N Natural Products Chemistry & Research ISSN: 2329-6836 Research Article Open Access Triterpenoid Saponins Garai S* CSIR-Indian Institute of Chemical Biology, 4 Raja SC Mullick Road, Jadavpur, Kolkata-700032, India Abstract A compilation of the 13C NMR data of the novel aglycones of new triterpenoid saponins, reported during the period mid1996-March, 2007 and arranged skeleton wise is included. The biological activities of the parent saponins are also covered. Keywords: Triterpenoid saponins; Isolation; Structure elucidation; Psolus patagonians was isolated as follows. The sea cucumbers, frozen Biological activity prior to storage were homogenized in EtOH and centrifuged. The dried ethanolic extract was partioned between MeOH-H2O (90:10) Introduction and cyclohexane. The methanalic extract was subjected to vacuum dry column chromatography on Davisil C reversed phase column (35- Triterpenoid saponins are naturally occurring surface active 18 70 μ) using H O and H O -MeOH mixtures. The fractions thus glycosides of triterpenes. They can be divided into two major groups a) 2 2 monodesmosides in which the aglycone has a singly attached linear or obtained were further purified by reverse phase HPLC togive the branched chain set of sugars and (b) bisdesmosides in which there pure saponin [11]. are two sets of sugars. They are widely distributed in plant kingdom, Structure elucidation certain marine organisms and marine flora and fauna. Their structural diversity, occurrence as complex mixtures, novel bioactivities of Structure elucidation of the isolated pure triterpenoid saponins is relevance to the pharmaceutical industry and agriculture, and usually carried out by a combination of chemical and spectroscopic usefulness as ingredients of cosmetics, allelochemicals, food methods. -

Steroidal Triterpenes of Cholesterol Synthesis
Molecules 2013, 18, 4002-4017; doi:10.3390/molecules18044002 OPEN ACCESS molecules ISSN 1420-3049 www.mdpi.com/journal/molecules Review Steroidal Triterpenes of Cholesterol Synthesis Jure Ačimovič and Damjana Rozman * Centre for Functional Genomics and Bio-Chips, Faculty of Medicine, Institute of Biochemistry, University of Ljubljana, Zaloška 4, Ljubljana SI-1000, Slovenia; E-Mail: [email protected] * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +386-1-543-7591; Fax: +386-1-543-7588. Received: 18 February 2013; in revised form: 19 March 2013 / Accepted: 27 March 2013 / Published: 4 April 2013 Abstract: Cholesterol synthesis is a ubiquitous and housekeeping metabolic pathway that leads to cholesterol, an essential structural component of mammalian cell membranes, required for proper membrane permeability and fluidity. The last part of the pathway involves steroidal triterpenes with cholestane ring structures. It starts by conversion of acyclic squalene into lanosterol, the first sterol intermediate of the pathway, followed by production of 20 structurally very similar steroidal triterpene molecules in over 11 complex enzyme reactions. Due to the structural similarities of sterol intermediates and the broad substrate specificity of the enzymes involved (especially sterol-Δ24-reductase; DHCR24) the exact sequence of the reactions between lanosterol and cholesterol remains undefined. This article reviews all hitherto known structures of post-squalene steroidal triterpenes of cholesterol synthesis, their biological roles and the enzymes responsible for their synthesis. Furthermore, it summarises kinetic parameters of enzymes (Vmax and Km) and sterol intermediate concentrations from various tissues. Due to the complexity of the post-squalene cholesterol synthesis pathway, future studies will require a comprehensive meta-analysis of the pathway to elucidate the exact reaction sequence in different tissues, physiological or disease conditions. -

Genetic Basis of Biosynthesis and Cytotoxic Activity of Medicago Truncatula Triterpene Saponins Brynn Kathleen Lawrence University of Arkansas, Fayetteville
University of Arkansas, Fayetteville ScholarWorks@UARK Theses and Dissertations 8-2016 Genetic Basis of Biosynthesis and Cytotoxic Activity of Medicago truncatula Triterpene Saponins Brynn Kathleen Lawrence University of Arkansas, Fayetteville Follow this and additional works at: http://scholarworks.uark.edu/etd Part of the Plant Biology Commons, Plant Breeding and Genetics Commons, and the Plant Pathology Commons Recommended Citation Lawrence, Brynn Kathleen, "Genetic Basis of Biosynthesis and Cytotoxic Activity of Medicago truncatula Triterpene Saponins" (2016). Theses and Dissertations. 1687. http://scholarworks.uark.edu/etd/1687 This Thesis is brought to you for free and open access by ScholarWorks@UARK. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of ScholarWorks@UARK. For more information, please contact [email protected], [email protected]. Genetic Basis of Biosynthesis and Cytotoxic Activity of Medicago truncatula Triterpene Saponins A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science in Cell and Molecular Biology By Brynn Kathleen Lawrence Ouachita Baptist University Bachelor of Science in Biology, 2014 August 2016 University of Arkansas This thesis is approved for recommendation to the Graduate Council. ______________________________________ Dr. Kenneth L. Korth Thesis Director ______________________________________ ______________________________________ Dr. Sun-Ok Lee Dr. David S. McNabb Committee Member Committee Member Abstract Saponins are a large family of specialized metabolites produced in many plants. They can have negative effects on a number of plant pests and are thought to play a role in plant defense. With current and possible future uses in industry and agriculture, saponins have also been shown to be hypocholesterolemic, hypoglycemic, immunostimulatory, antioxidative, anti-inflammatory, and cytotoxic. -

Expression of Selected Genes Involved in Steroidogenesis in the Course of Enucleation-Induced Rat Adrenal Regeneration
INTERNATIONAL JOURNAL OF MOLECULAR MEDICINE 33: 613-623, 2014 Expression of selected genes involved in steroidogenesis in the course of enucleation-induced rat adrenal regeneration MARIANNA TYCZEWSKA, MARCIN RUCINSKI, AGNIESZKA ZIOLKOWSKA, MARCIN TREJTER, MARTA SZYSZKA and LUDWIK K. MALENDOWICZ Department of Histology and Embryology, Poznan University of Medical Sciences, Poznan, Poland Received October 28, 2013; Accepted December 6, 2013 DOI: 10.3892/ijmm.2013.1599 Abstract. The enucleation-induced (EI) rapid proliferation however, throughout the entire experimental period, there were of adrenocortical cells is followed by their differentiation, no statistically significant differences observed. After the initial the degree of which may be characterized by the expression decrease in steroidogenic factor 1 (Sf-1) mRNA levels observed of genes directly and indirectly involved in steroid hormone on the 1st day of the experiment, a marked upregulation in its biosynthesis. In this study, out of 30,000 transcripts of genes expression was observed from there on. Data from the current identified by means of Affymetrix Rat Gene 1.1 ST Array, we study strongly suggest the role of Fabp6, Lipe and Soat1 in aimed to select genes (either up- or downregulated) involved supplying substrates of regenerating adrenocortical cells for in steroidogenesis in the course of enucleation-induced adrenal steroid synthesis. Our results indicate that during the first days regeneration. On day 1, we found 32 genes with altered of adrenal regeneration, intense synthesis of cholesterol may expression levels, 15 were upregulated and 17 were down- occur, which is then followed by its conversion into cholesteryl regulated [i.e., 3β-hydroxysteroid dehydrogenase (Hsd3β), esters. -
Figures Intermediary Ontogeny.Pptx
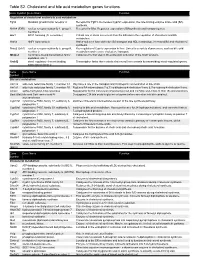
Table S2. Cholesterol and bile acid metabolism genes functions. Gene Symbol Gene Name Function Regulation of cholesterol and/or bile acid metabolism Fgfr4 fibroblast growth factor receptor 4 Receptor for Fgf15. Decreases Cyp7a1 expression, the rate-limiting enzyme in bile acid (BA) synthesis. Nr1h4 (FXR) nuclear receptor subfamily 1, group H, Receptor for BAs. Regulates expression of BAsynthesis and transport genes. member 4 Arv1 ARV1 homolog (S. cerevisiae) Critical role in sterol movement from the ER and in the regulation of cholesterol and BA metabolism. Hnf1a HNF1 homeobox A Hnf1a-null mice have defective BA transport and HDL metabolism, increased BA and cholesterol synthesis. Nr5a2 (Lrh1) nuclear receptor subfamily 5, group A, Key regulator of Cyp7a expression in liver. Linked to a variety of processes, such as bile acid member 2 metabolism and reverse cholesterol transport. Mbtps1 membrane-bound transcription factor Catalyzes the first step in the proteolytic activation of the Srebf proteins. peptidase, site 1 Srebf2 sterol regulatory element binding Transcription factor that controls cholesterol homeostasis by transcribing sterol-regulated genes. transcription factor 2 Gene Gene Name Function Symbol Bile acid metabolism Akr1c6 aldo-keto reductase family 1, member C1 May have a role in the transport and intrahepatic concentration of bile acids. Akr1d1 aldo-keto reductase family 1, member D1 Reduces BA intermediates 7-α,12-α-dihydroxy-4-cholesten-3-one & 7-α-hydroxy-4-cholesten-3-one. Amacr alpha-methylacyl-CoA racemase Responsible for the conversion of pristanoyl-CoA and C27-bile acyl-CoAs to their (S)-stereoisomers. Baat (Bat) bile acid CoA: amino acid N- Conjugates C24 bile acids to glycine or taurine before excretion into bile canaliculi. -

GRADUATE INSTITUTE of HEALTH SCIENCE TRITERPENE SAPONINS from the SEED COAT of QUINOA (Chenopodium Quinoa Willd.) Farman Ullah K
GRADUATE INSTITUTE OF HEALTH SCIENCE TRITERPENE SAPONINS FROM THE SEED COAT OF QUINOA (Chenopodium Quinoa Willd.) Farman Ullah KHAN PHARMACOGNOSY MASTER THESIS Nicosia 2019 GRADUATE INSTITUTE OF HEALTH SCIENCE TRITERPENE SAPONINS FROM THE SEED COAT OF QUINOA (Chenopodium Quinoa Willd.) Farman Ullah KHAN PHARMACOGNOSY MASTER THESIS SUPERVISOR Prof. Dr. İhsan ÇALIŞ Co-SUPERVISOR Prof. Dr. K. Hüsnü C. BAŞER Lefkoşa (Nicosia) 2019 TABLE OF CONTENTS CONTENT PAGE ÖZET III ABSTRACT IV FIGURES V PICTURES V SCHEMES V SPECTRA VI TABLES VII 1. INTRODUCTION 1 2. LITERATURE REVIEW 2 2.1. Botanical Characters 3 2.1.1. Amaranthaceae Family 3 2.1.1.1. Distribution 4 2.1.1.2. Chenopodium quinoa 4 2.2. Phytochemistry and Nutritional Value of Quinoa 7 2.2.1. Saponins 7 2.2.2. The Other Secondary Metabolites 8 2.2.3. Quinoa and Gluten 13 2.2.4. Vitamins 13 2.2.5. Minerals 14 2.2.6. Carbohydrate and fiber 15 2.2.7. Proteins 16 2.3. Pharmacological Activities 17 2.3.1. Antioxidant Activities 17 2.3.1.1. Antioxidant and anticancer activities of Chenopodium quinoa 17 leaves extracts 2.3.2. Antimicrobial Activities 17 2.3.3. Antienflammatuar Activities 18 2.3.4. Anti-obesity and Antidiabetic Activities 19 I Page 2.3.5. Other Activities 20 3. EXPERIMENTAL PART 21 3.1. Plant Material 21 3.2. Material and Methods 21 3.2.1. Chemical Solid Materials 21 3.2.2. Chromatographic Methods 21 3.2.2.1. Thin Layer Chromatography 21 3.2.2.2. Vacuum Column Chromatography (VCC) 22 3.2.2.3.