Evaluation of the Relatedness of Brucella Spp. and Ochrobactrum Anthropi and Description of Ochrobactrum Intermedium Sp
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Iron Transport Strategies of the Genus Burkholderia
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2015 Iron transport strategies of the genus Burkholderia Mathew, Anugraha Posted at the Zurich Open Repository and Archive, University of Zurich ZORA URL: https://doi.org/10.5167/uzh-113412 Dissertation Published Version Originally published at: Mathew, Anugraha. Iron transport strategies of the genus Burkholderia. 2015, University of Zurich, Faculty of Science. Iron transport strategies of the genus Burkholderia Dissertation zur Erlangung der naturwissenschaftlichen Doktorwürde (Dr. sc. nat.) vorgelegt der Mathematisch-naturwissenschaftlichen Fakultät der Universität Zürich von Anugraha Mathew aus Indien Promotionskomitee Prof. Dr. Leo Eberl (Vorsitz) Prof. Dr. Jakob Pernthaler Dr. Aurelien carlier Zürich, 2015 2 Table of Contents Summary .............................................................................................................. 7 Zusammenfassung ................................................................................................ 9 Abbreviations ..................................................................................................... 11 Chapter 1: Introduction ....................................................................................... 14 1.1.Role and properties of iron in bacteria ...................................................................... 14 1.2.Iron transport mechanisms in bacteria ..................................................................... -

Appendix a Bacteria
Appendix A Complete list of 594 pathogens identified in canines categorized by the following taxonomical groups: bacteria, ectoparasites, fungi, helminths, protozoa, rickettsia and viruses. Pathogens categorized as zoonotic/sapronotic/anthroponotic have been bolded; sapronoses are specifically denoted by a ❖. If the dog is involved in transmission, maintenance or detection of the pathogen it has been further underlined. Of these, if the pathogen is reported in dogs in Canada (Tier 1) it has been denoted by an *. If the pathogen is reported in Canada but canine-specific reports are lacking (Tier 2) it is marked with a C (see also Appendix C). Finally, if the pathogen has the potential to occur in Canada (Tier 3) it is marked by a D (see also Appendix D). Bacteria Brachyspira canis Enterococcus casseliflavus Acholeplasma laidlawii Brachyspira intermedia Enterococcus faecalis C Acinetobacter baumannii Brachyspira pilosicoli C Enterococcus faecium* Actinobacillus Brachyspira pulli Enterococcus gallinarum C C Brevibacterium spp. Enterococcus hirae actinomycetemcomitans D Actinobacillus lignieresii Brucella abortus Enterococcus malodoratus Actinomyces bovis Brucella canis* Enterococcus spp.* Actinomyces bowdenii Brucella suis Erysipelothrix rhusiopathiae C Actinomyces canis Burkholderia mallei Erysipelothrix tonsillarum Actinomyces catuli Burkholderia pseudomallei❖ serovar 7 Actinomyces coleocanis Campylobacter coli* Escherichia coli (EHEC, EPEC, Actinomyces hordeovulneris Campylobacter gracilis AIEC, UPEC, NTEC, Actinomyces hyovaginalis Campylobacter -
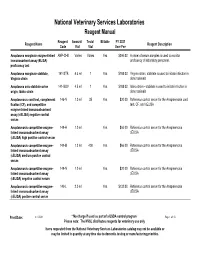
NVSL Reagent Manual
National Veterinary Services Laboratories Reagent Manual Reagent Amount/ Tests/ Billable FY 2021 Reagent Name Reagent Description Code Vial Vial User Fee Anaplasma marginale enzyme-linked ANP-CHK Varies VariesYes $394.00 A panel of serum samples is used to monitor immunosorbent assay (ELISA) proficiency of laboratory personnel. proficiency test Anaplasma marginale stabilate, 141-STB 4.5 ml 1Yes $188.00 Virginia strain, stabilate is used to initiate infection in Virginia strain donor animals Anaplasma ovis stabilate ovine 141-SOV 4.5 ml 1Yes $188.00 Idaho strain - stabilate is used to initate infection in origin, Idaho strain donor animals Anaplasmosis card test, complement 146-N 1.0 ml 35Yes $20.00 Reference control serum for the Anaplasmosis card fixation (CF), and competitive test, CF, and cELISA enzyme-linked immunoabsorbent assay (cELISA) negative control serum Anaplasmosis competitive enzyme- 149-H 1.0 mlYes $66.00 Reference control serum for the Anaplasmosis linked immunoabsorbent assay cELISA (cELISA) high positive control serum Anaplasmosis competitive enzyme- 149-M 1.0 ml 400Yes $66.00 Reference control serum for the Anaplasmosis linked immunoabsorbent assay cELISA (cELISA) medium positve control serum Anaplasmosis competitive enzyme- 149-N 1.0 mlYes $20.00 Reference control serum for the Anaplasmosis linked immunoabsorbent assay cELISA (cELISA) negative control serum Anaplasmosis competitive enzyme- 149-L 2.0 mlYes $132.00 Reference control serum for the Anaplasmosis linked immunoabsorbent assay cELISA (cELISA) positve control serum Print Date: 6/11/2021 * No charge if used as part of a USDA control program Page 1 of 58 Please note: The NVSL distributes reagents for veterinary use only Items requested from the National Veterinary Services Laboratories catalog may not be available or may be limited in quantity at any time due to domestic testing or manufacturing priorities. -

APUTS) Reporting Terminology and Codes Microbiology (V1.0
AUSTRALIAN PATHOLOGY UNITS AND TERMINOLOGY (APUTS) Reporting Terminology and Codes Microbiology (v1.0) 1 12/02/2013 APUTS Report Information Model - Urine Microbiology Page 1 of 1 Specimen Type Specimen Macro Time Glucose Bilirubin Ketones Specific Gravity pH Chemistry Protein Urobilinogen Nitrites Haemoglobin Leucocyte Esterases White blood cell count Red blood cells Cells Epithelial cells Bacteria Microscopy Parasites Microorganisms Yeasts Casts Crystals Other elements Antibacterial Activity No growth Mixed growth Urine MCS No significant growth Klebsiella sp. Bacteria ESBL Klebsiella pneumoniae Identification Virus Fungi Growth of >10^8 org/L 10^7 to 10^8 organism/L of mixed Range or number Colony Count growth of 3 organisms 19090-0 Culture Organism 1 630-4 LOINC >10^8 organisms/L LOINC Significant growth e.g. Ampicillin 18864-9 LOINC Antibiotics Susceptibility Method Released/suppressed None Organism 2 Organism 3 Organism 4 None Consistent with UTI Probable contamination Growth unlikely to be significant Comment Please submit a repeat specimen for testing if clinically indicated Catheter comments Sterile pyuria Notification to infection control and public health departments PUTS Urine Microbiology Information Model v1.mmap - 12/02/2013 - Mindjet 12/02/2013 APUTS Report Terminology and Codes - Microbiology - Urine Page 1 of 3 RCPA Pathology Units and Terminology Standardisation Project - Terminology for Reporting Pathology: Microbiology : Urine Microbiology Report v1 LOINC LOINC LOINC LOINC LOINC LOINC LOINC Urine Microbiology Report -

Characterizing the Abcr/Vtlr System in the Rhizobiales
Characterizing the AbcR/VtlR system in the Rhizobiales Lauren Marie Sheehan Dissertation submitted to the faculty of the Virginia Polytechnic Institute and State University in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical and Veterinary Sciences Clayton C. Caswell, Chair Thomas J. Inzana Birgit Scharf Nammalwar Sriranganathan April 20, 2018 Blacksburg, Virginia Keywords: Brucella, LysR-type transcriptional regulators, small RNAs Characterizing the AbcR/VtlR system in the Rhizobiales Lauren Marie Sheehan Abstract Rhizobiales encompass a diverse group of microbes, ranging from free-living, soil- dwelling bacteria to disease-causing, intracellular pathogens. Although the lifestyle of these organisms vary, many genetic systems are well conserved. One system, named the AbcR/VtlR system, is found throughout rhizobiales, and even extends to bacteria in other orders within the Alphaproteobacteria. The AbcR sRNAs are an example of sibling sRNAs, where two copies of the abcR gene are typically present in the genome. The AbcRs are involved in the negative regulation of ABC-type transport systems, which are important components for nutrient acquisition. Although the AbcRs share several features amongst organisms, major differences can be found in their functional and regulatory redundancy, the targets they regulate and how they regulate them. Specifically, one major difference in the AbcRs lies in the nucleotide sequences utilized by the sRNAs to bind mRNA targets. In the present studies, the regulatory mechanisms of the AbcR sRNAs were further characterized in the mammalian pathogen Brucella abortus, and the full regulatory profiles of the AbcRs were defined in the plant pathogen Agrobacterium tumefaciens. As mentioned above, the AbcR sRNAs are important for the proper regulation of nutrient-acquiring transport systems in the Rhizobiales. -

Tese Adelina Margarida Parente
Defences of Helicobacter species against host antimicrobials Adelina Margarida Lima Pereira Rodrigues Parente Dissertation presented to obtain the Ph.D degree in Biochemistry Instituto de Tecnologia Química e Biológica António Xavier | Universidade Nova de Lisboa Oeiras, June, 2016 Defences of Helicobacter species against host antimicrobials Adelina Margarida Lima Pereira Rodrigues Parente Dissertation presented to obtain the Ph.D degree in Biochemistry Instituto de Tecnologia Química e Biológica António Xavier | Universidade Nova de Lisboa Oeiras, June 2016 From left to right: Mónica Oleastro (4th oponente), Gabriel Martins (3 rd opponent), Marta Justino (Co-supervisor) , Miguel Viveiros (2 nd opponent), Adelina Margarida Parente , Lígia Saraiva (supervisor) , Cecília Arraiano (president of the jury), and Maria do Céu Figueiredo (1 st opponent). nd 22 June 2016 Second edition, June 2016 Molecular Mechanisms of Pathogen Resistance Laboratory Instituto de Tecnologia Química e Biológica António Xavier Universidade Nova de Lisboa 2780-157 Portugal ii “Science knows no country, because knowledge belongs to humanity, and is the torch which illuminates the world” Louis Pasteur iii iv Acknowledgments Firstly, I would like to express my gratitude to the person that allowed me the opportunity to perform a PhD and who also contributed the most for my accomplishment of this thesis by constantly supporting me during these last years. I thus thank my supervisor, Dr. Lígia Saraiva , for her permanent availability whenever I needed guidance, for all the excellent ideas and advices related to my practical work and lastly for all the patience and enthusiasm! I also thank Dr. Lígia for her rigour and enormous help in the writing of this thesis. -

An Oxygen-Sensing Two-Component System in the Burkholderia Cepacia Complex Regulates Biofilm, Intracellular Invasion, and Pathogenicity
An Oxygen-Sensing Two-Component System in the Burkholderia cepacia Complex Regulates Biofilm, Intracellular Invasion, and Pathogenicity The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Schaefers, Matthew M., Tiffany L. Liao, Nicole M. Boisvert, Damien Roux, Deborah Yoder-Himes, and Gregory P. Priebe. 2017. “An Oxygen-Sensing Two-Component System in the Burkholderia cepacia Complex Regulates Biofilm, Intracellular Invasion, and Pathogenicity.” PLoS Pathogens 13 (1): e1006116. doi:10.1371/journal.ppat.1006116. http://dx.doi.org/10.1371/ journal.ppat.1006116. Published Version doi:10.1371/journal.ppat.1006116 Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:30371001 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA RESEARCH ARTICLE An Oxygen-Sensing Two-Component System in the Burkholderia cepacia Complex Regulates Biofilm, Intracellular Invasion, and Pathogenicity Matthew M. Schaefers1,2*, Tiffany L. Liao1, Nicole M. Boisvert1, Damien Roux3, Deborah Yoder-Himes4, Gregory P. Priebe1,2 a1111111111 1 Division of Critical Care Medicine, Department of Anesthesiology, Perioperative and Pain Medicine, Boston Children's Hospital, Boston, Massachusetts, United States of America, 2 Department of Anaesthesia, a1111111111 Harvard Medical School, Boston, -

Highly Sensitive Bacteriophage-Based Detection of Brucella Abortus in Mixed Culture and Spiked Blood
Article Highly Sensitive Bacteriophage-Based Detection of Brucella abortus in Mixed Culture and Spiked Blood Kirill V. Sergueev, Andrey A. Filippov and Mikeljon P. Nikolich * Department of Bacteriophage Therapeutics, Bacterial Diseases Branch, Walter Reed Army Institute of Research, Silver Spring, MD 20910, USA; [email protected] (K.V.S.); [email protected] (A.A.F.) * Correspondence: [email protected]; Tel.: +1-301-319-9469 Academic Editor: Tessa E. F. Quax, Matthias G. Fischer and Laurent Debarbieux Received: 1 April 2017; Accepted: 6 June 2017; Published: 10 June 2017 Abstract: For decades, bacteriophages (phages) have been used for Brucella species identification in the diagnosis and epidemiology of brucellosis. Traditional Brucella phage typing is a multi-day procedure including the isolation of a pure culture, a step that can take up to three weeks. In this study, we focused on the use of brucellaphages for sensitive detection of the pathogen in clinical and other complex samples, and developed an indirect method of Brucella detection using real-time quantitative PCR monitoring of brucellaphage DNA amplification via replication on live Brucella cells. This assay allowed the detection of single bacteria (down to 1 colony-forming unit per milliliter) within 72 h without DNA extraction and purification steps. The technique was equally efficient with Brucella abortus pure culture and with mixed cultures of B. abortus and α- proteobacterial near neighbors that can be misidentified as Brucella spp., Ochrobactrum anthropi and Afipia felis. The addition of a simple short sample preparation step enabled the indirect phage-based detection of B. -

The Intracellular Behaviour of Burkholderia Cenocepacia in Murine Macrophages
Western University Scholarship@Western Electronic Thesis and Dissertation Repository 11-28-2011 12:00 AM The intracellular behaviour of Burkholderia cenocepacia in murine macrophages Jennifer S. Tolman The University of Western Ontario Supervisor Dr. Miguel A. Valvano The University of Western Ontario Graduate Program in Microbiology and Immunology A thesis submitted in partial fulfillment of the equirr ements for the degree in Doctor of Philosophy © Jennifer S. Tolman 2011 Follow this and additional works at: https://ir.lib.uwo.ca/etd Part of the Cell Biology Commons, Genetics Commons, and the Microbiology Commons Recommended Citation Tolman, Jennifer S., "The intracellular behaviour of Burkholderia cenocepacia in murine macrophages" (2011). Electronic Thesis and Dissertation Repository. 338. https://ir.lib.uwo.ca/etd/338 This Dissertation/Thesis is brought to you for free and open access by Scholarship@Western. It has been accepted for inclusion in Electronic Thesis and Dissertation Repository by an authorized administrator of Scholarship@Western. For more information, please contact [email protected]. THE INTRACELLULAR BEHAVIOUR OF BURKHOLDERIA CENOCEPACIA IN MURINE MACROPHAGES (Spine title: Intramacrophage behaviour of B. cenocepacia) (Thesis format: Monograph) by Jennifer Sarah Tolman Graduate Program in Microbiology and Immunology A thesis submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy The School of Graduate and Postdoctoral Studies The University of Western Ontario London, Ontario, Canada © Jennifer S. Tolman 2012 THE UNIVERSITY OF WESTERN ONTARIO School of Graduate and Postdoctoral Studies CERTIFICATE OF EXAMINATION Supervisor Examiners ______________________________ ______________________________ Dr. Miguel Valvano Dr. France Daigle Supervisory Committee ______________________________ Dr. Charles Trick ______________________________ Dr. John McCormick ______________________________ Dr. -

Brucellosis in Humans and Livestock in Rural Uganda: Epidemiology and Tools for Resource-Limited Settings
BRUCELLOSIS IN HUMANS AND LIVESTOCK IN RURAL UGANDA: EPIDEMIOLOGY AND TOOLS FOR RESOURCE-LIMITED SETTINGS By RoseAnn Miller A DISSERTATION Submitted to Michigan State University in partial fulfillment of the requirements for the degree of Large Animal Clinical Sciences – Doctor of Philosophy 2016 ABSTRACT BRUCELLOSIS IN HUMANS AND LIVESTOCK IN RURAL UGANDA: EPIDEMIOLOGY AND TOOLS FOR RESOURCE-LIMITED SETTINGS By RoseAnn Miller Objectives: A cross-sectional study was undertaken to describe the epidemiology of zoonotic brucellosis in dairy farms in rural southwestern Uganda, and to evaluate the use of tools to address challenges to disease surveillance programs faced in rural sub-Saharan Africa and other resource-limited settings. Methods: Blood samples were collected from cattle, goats and humans, and milk samples were collected from cows on 70 dairy farms in two sites in Uganda. Samples of blood and milk from livestock were collected and dried on laboratory-grade filter paper. Data describing each animal and human subject were recorded during sample collection, and herd-level risk factor data were collected by questionnaires on livestock health and management, human health and practices associated with increased brucellosis risk. Livestock blood and milk samples were tested using the Rose Bengal (RBT) and milk ring (MRT) tests. Human blood was tested using an immunochromatographic lateral flow assay (LFA). A qualitative real-time PCR (q-PCR) and nested PCR (n-PCR) were used to detect Brucella in DNA extracted from human, cattle and goat blood clots, cow milk, and human sera, and q-PCR was used to test DNA extracted from cattle and goat dried blood and milk samples. -

The Epidemiology of Brucellosis in Sheep, Goats and Humans in the Iraqi Kurdistan Region
The Epidemiology of Brucellosis in Sheep, Goats and Humans in the Iraqi Kurdistan Region Emad Abdlghafoor Aziz Alshwany (BVMS, HDipVM/Poultry Diseases & MVS) A thesis presented in fulfilment of the requirements for the degree of Doctor of Philosophy School of Veterinary Medicine College of Sciences, Health, Engineering and Education Murdoch University Western Australia 2019 Declaration I declare that this thesis is my own account of my research and contains as its main content work which has not previously been submitted for a degree at any tertiary education institution. Emad Alshwany ii Dedication I dedicate this Doctoral Thesis to the most precious people in my life, for their everlasting love and strong support in my study: My parents, wife, daughters, son, sister and brothers. iii Abstract Brucellosis is a disease affecting a wide range of domesticated animals and wildlife as well as humans. The disease remains a major zoonotic problem in many regions including the Middle East. In Iraq, where brucellosis is endemic, the disease is a major economic and production limiting disease for livestock owners and the community. Impacts on production arise from reduced milk production, abortions, decreased reproduction rate and premature births. The aim of this project was to investigate the seroprevalence, risk factors and economic impact of brucellosis in sheep and goats in the Kurdistan Region. Also a retrospective study of human brucellosis in Iraq was conducted to describe the historical distribution of the disease and its impact on the population. Fifty one (39 sheep and 12 goats) of 1,050 sera samples were positive on both an RBT and ELISA (overall seroprevalence of 4.9%; 95%CI 3.6 - 6.3). -

G1-Arrested Newborn Cells Are the Predominant Infectious Form of the Pathogen Brucella Abortus
ARTICLE Received 25 Oct 2013 | Accepted 10 Jun 2014 | Published 9 Jul 2014 DOI: 10.1038/ncomms5366 OPEN G1-arrested newborn cells are the predominant infectious form of the pathogen Brucella abortus Michae¨l Deghelt1,*, Caroline Mullier1,*, Jean- Franc¸ois Sternon1, Nayla Francis1,Ge´raldine Laloux2,3,4, Delphine Dotreppe1, Charles Van der Henst1, Christine Jacobs-Wagner2,3,4, Jean-Jacques Letesson1 & Xavier De Bolle1 Several intracellular pathogens, such as Brucella abortus, display a biphasic infection process starting with a non-proliferative stage of unclear nature. Here, we study the cell cycle of B. abortus at the single-cell level, in culture and during infection of HeLa cells and macrophages. The localization of segregation and replication loci of the two bacterial chromosomes indicates that, immediately after being engulfed by host-cell endocytic vacuoles, most bacterial cells are newborn. These bacterial cells do not initiate DNA replication for the next 4 to 6 h, indicating a G1 arrest. Moreover, growth is completely stopped during that time, reflecting a global cell cycle block. Growth and DNA replication resume later, although bacteria still reside within endosomal-like compartments. We hypothesize that the predominance of G1-arrested bacteria in the infectious population, and the bacterial cell cycle arrest following internalization, may constitute a widespread strategy among intracellular pathogens to colonize new proliferation niches. 1 Microorganisms biology research unit (URBM), University of Namur (UNamur), Namur, Belgium. 2 Department of Molecular, Cellular and Developmental Biology, Yale University, New Haven, Connecticut 06520, USA. 3 Howard Hughes Medical Institute, Yale University, New Haven, Connecticut 06520, USA. 4 Department of Microbial Pathogenesis, Yale School of Medicine, New Haven, Connecticut 06510, USA.