Rangewide Tidewater Goby Occupancy Survey Using Environmental Dna
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

San Francisco Bay Area Integrated Regional Water Management Plan
San Francisco Bay Area Integrated Regional Water Management Plan October 2019 Table of Contents List of Tables ............................................................................................................................... ii List of Figures.............................................................................................................................. ii Chapter 1: Governance ............................................................................... 1-1 1.1 Background ....................................................................................... 1-1 1.2 Governance Team and Structure ...................................................... 1-1 1.2.1 Coordinating Committee ......................................................... 1-2 1.2.2 Stakeholders .......................................................................... 1-3 1.2.2.1 Identification of Stakeholder Types ....................... 1-4 1.2.3 Letter of Mutual Understandings Signatories .......................... 1-6 1.2.3.1 Alameda County Water District ............................. 1-6 1.2.3.2 Association of Bay Area Governments ................. 1-6 1.2.3.3 Bay Area Clean Water Agencies .......................... 1-6 1.2.3.4 Bay Area Water Supply and Conservation Agency ................................................................. 1-8 1.2.3.5 Contra Costa County Flood Control and Water Conservation District .................................. 1-8 1.2.3.6 Contra Costa Water District .................................. 1-9 1.2.3.7 -

Marin Islands NWR Sport Fishing Plan
Table of Contents Table of Contents 2 MARIN ISLANDS NATIONAL WILDLIFE REFUGE 3 SPORT FISHING PLAN 3 1. Introduction 3 2. Statement of Objectives 4 3. Description of the Fishing Program 5 A. Area to be Opened to Fishing 5 B. Species to be Taken, Fishing periods, Fishing Access 5 C. Fishing Permit Requirements 7 D. Consultation and Coordination with the State 7 E. Law Enforcement 7 F. Funding and Staffing Requirements 8 4. Conduct of the Fishing Program 8 A. Permit Application, Selection, and/or Registration Procedures (if applicable) 8 B. Refuge-Spec if ic Fishing Regulat ions 8 C. Relevant State Regulations 8 D. Other Refuge Rules and Regulations for Sport Fishing 8 5. Public Engagement 9 A. Outreach for Announcing and Publicizing the Fishing Program 9 B. Anticipated Public Reaction to the Fishing Program 9 C. How Anglers Will Be Informed of Relevant Rules and Regulations 9 6. Compatibility Determination 9 7. Literature Cited 9 List of Figures Figure 1. Proposed Sport Fishing Area Fishing…………………………………6 Marin Islands NWR Fishing Plan Page 2 MARIN ISLANDS NATIONAL WILDLIFE REFUGE SPORT FISHING PLAN 1. Introduction National Wildlife Refuges are guided by the mission and goals of the National Wildlife Refuge System (NWRS), the purposes of an individual refuge, Service policy, and laws and international treaties. Relevant guidance includes the National Wildlife Refuge System Administration Act of 1966, as amended by the National Wildlife Refuge System Improvement Act of 1997, Refuge Recreation Act of 1962, and selected portions of the Code of Federal Regulations and Fish and Wildlife Service Manual. -

Section 3.4 Biological Resources 3.4- Biological Resources
SECTION 3.4 BIOLOGICAL RESOURCES 3.4- BIOLOGICAL RESOURCES 3.4 BIOLOGICAL RESOURCES This section discusses the existing sensitive biological resources of the San Francisco Bay Estuary (the Estuary) that could be affected by project-related construction and locally increased levels of boating use, identifies potential impacts to those resources, and recommends mitigation strategies to reduce or eliminate those impacts. The Initial Study for this project identified potentially significant impacts on shorebirds and rafting waterbirds, marine mammals (harbor seals), and wetlands habitats and species. The potential for spread of invasive species also was identified as a possible impact. 3.4.1 BIOLOGICAL RESOURCES SETTING HABITATS WITHIN AND AROUND SAN FRANCISCO ESTUARY The vegetation and wildlife of bayland environments varies among geographic subregions in the bay (Figure 3.4-1), and also with the predominant land uses: urban (commercial, residential, industrial/port), urban/wildland interface, rural, and agricultural. For the purposes of discussion of biological resources, the Estuary is divided into Suisun Bay, San Pablo Bay, Central San Francisco Bay, and South San Francisco Bay (See Figure 3.4-2). The general landscape structure of the Estuary’s vegetation and habitats within the geographic scope of the WT is described below. URBAN SHORELINES Urban shorelines in the San Francisco Estuary are generally formed by artificial fill and structures armored with revetments, seawalls, rip-rap, pilings, and other structures. Waterways and embayments adjacent to urban shores are often dredged. With some important exceptions, tidal wetland vegetation and habitats adjacent to urban shores are often formed on steep slopes, and are relatively recently formed (historic infilled sediment) in narrow strips. -
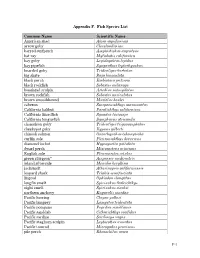
Appendix E: Fish Species List
Appendix F. Fish Species List Common Name Scientific Name American shad Alosa sapidissima arrow goby Clevelandia ios barred surfperch Amphistichus argenteus bat ray Myliobatis californica bay goby Lepidogobius lepidus bay pipefish Syngnathus leptorhynchus bearded goby Tridentiger barbatus big skate Raja binoculata black perch Embiotoca jacksoni black rockfish Sebastes melanops bonehead sculpin Artedius notospilotus brown rockfish Sebastes auriculatus brown smoothhound Mustelus henlei cabezon Scorpaenichthys marmoratus California halibut Paralichthys californicus California lizardfish Synodus lucioceps California tonguefish Symphurus atricauda chameleon goby Tridentiger trigonocephalus cheekspot goby Ilypnus gilberti chinook salmon Oncorhynchus tshawytscha curlfin sole Pleuronichthys decurrens diamond turbot Hypsopsetta guttulata dwarf perch Micrometrus minimus English sole Pleuronectes vetulus green sturgeon* Acipenser medirostris inland silverside Menidia beryllina jacksmelt Atherinopsis californiensis leopard shark Triakis semifasciata lingcod Ophiodon elongatus longfin smelt Spirinchus thaleichthys night smelt Spirinchus starksi northern anchovy Engraulis mordax Pacific herring Clupea pallasi Pacific lamprey Lampetra tridentata Pacific pompano Peprilus simillimus Pacific sanddab Citharichthys sordidus Pacific sardine Sardinops sagax Pacific staghorn sculpin Leptocottus armatus Pacific tomcod Microgadus proximus pile perch Rhacochilus vacca F-1 plainfin midshipman Porichthys notatus rainwater killifish Lucania parva river lamprey Lampetra -

Report on the Monitoring of Radionuclides in Fishery Products (March 2011 - January 2015)
Report on the Monitoring of Radionuclides in Fishery Products (March 2011 - January 2015) April 2015 Fisheries Agency of Japan 0 1 Table of Contents Overview…………………………………………………………………………………………………. 8 The Purpose of this Report………………………………………………………………………………9 Part One. Efforts to Guarantee the Safety of Fishery Products………………………………………..11 Chapter 1. Monitoring of Radioactive Materials in Food; Restrictions on Distribution and Other Countermeasures………...…………………………………………………………………11 1-1-1 Standard Limits for Radioactive Materials in Food………………………………………...……11 1-1-2 Methods of Testing for Radioactive Materials………………………………………...…………12 1-1-3 Inspections of Fishery Products for Radioactive Materials…………………………...…………14 1-1-4 Restrictions and Suspensions on Distribution and Shipping ……………………………………..18 1-1-5 Cancellation of Restrictions on Shipping and Distribution………………………………………20 Box 1 Calculation of the Limits for Human Consumption……..………………………………………23 Box 2 Survey of Radiation Dose from Radionuclides in Foods Calculation of the Limits…………….24 Box 3 Examples of Local Government Monitoring Plan………………………………...…………….25 Chapter 2. Results of Radioactive Cesium Inspections for Fishery Products…………………………26 1-2-1 Inspection Results for Nationwide Fishery Products in Japan (in total)…………………………26 1-2-2 Inspection Results for Fukushima Prefecture Fishery Products (all)…………………………….27 1-2-3 Inspection Results for Fishery Products (all) from Outside Fukushima Prefecture……………...30 1-2-4 Trends within Fish Species……………………………………………………………………….32 1-2-5 Inspection Results for Main Target Fish Species of Fishing and Farming by Fiscal Year……….42 1-2-6 Radioactive Material Concentrations within Fish within 20 km of the Fukushima Daiichi NPS.46 Box 4 Fukushima Fishing Trials………………………………...……………………………………...47 1-2-7 Screening Test by Prefectural and Municipal Governments……………………………………..48 Chapter 3. Inspection for Radionuclides Other Than Radioactive Cesium……………………………49 1-3-1 Inspections for Radioactive Strontium etc. -

A Dissertation Entitled Evolution, Systematics
A Dissertation Entitled Evolution, systematics, and phylogeography of Ponto-Caspian gobies (Benthophilinae: Gobiidae: Teleostei) By Matthew E. Neilson Submitted as partial fulfillment of the requirements for The Doctor of Philosophy Degree in Biology (Ecology) ____________________________________ Adviser: Dr. Carol A. Stepien ____________________________________ Committee Member: Dr. Christine M. Mayer ____________________________________ Committee Member: Dr. Elliot J. Tramer ____________________________________ Committee Member: Dr. David J. Jude ____________________________________ Committee Member: Dr. Juan L. Bouzat ____________________________________ College of Graduate Studies The University of Toledo December 2009 Copyright © 2009 This document is copyrighted material. Under copyright law, no parts of this document may be reproduced without the expressed permission of the author. _______________________________________________________________________ An Abstract of Evolution, systematics, and phylogeography of Ponto-Caspian gobies (Benthophilinae: Gobiidae: Teleostei) Matthew E. Neilson Submitted as partial fulfillment of the requirements for The Doctor of Philosophy Degree in Biology (Ecology) The University of Toledo December 2009 The study of biodiversity, at multiple hierarchical levels, provides insight into the evolutionary history of taxa and provides a framework for understanding patterns in ecology. This is especially poignant in invasion biology, where the prevalence of invasiveness in certain taxonomic groups could -

Inland Fishes of California
Inland Fishes of California Revise d and Expanded PETER B. MO YL E Illustrations by Chris Ma ri van Dyck and Joe Tome ller i NIVERS ITyor ALfFORNJA PRESS Ikrkd cr I.", ..\ n~d e ' Lon don Universit }, 0 Ca lifornia Press Herkdey and Los Angeles, Ca lifornia Uni ve rsity of alifornia Press, Ltd. Lundun, England ~ 2002 by the Regents of the Unive rsi ty of Ca lifornia Library of Cungress ataloging-in -Publ ica tion Data j\·[oyk, Pen: r B. Inland fis hes of California / Peter B. Moyle ; illustrations by Chris Mari van D)'ck and Joe Tomell eri.- Rev. and expanded. p. cm. In lu de> bibl iographical refe rences (p. ). ISBN 0- 20-2.2754 -'1 (cl ot h: alk. papa) I. rreshw:ltcr lishes-Cali(ornia. I. Title. QL62S C2 M6H 2002 597 .17/i'097Q4-dc21 20010 27680 1\!UlIl.Ifaclu rcd in Canacla II 10 Q9 00 07 06 0 04 03 02 10 ' 1\ 7 b '; -\ 3 2 1 Th paper u!)ed in thi> public.ltiu(] 111l'd., the minimum requirements "fA SI / i': ISO Z39.4H-1992 (R 199;) ( Pmlllllh'/l e ofPa pcr) . e Special Thanks The illustrations for this book were made possible by gra nts from the following : California-Nevada Chapter, American Fi she ries Soc iety Western Di vision, Am erican Fi she ries Society California Department of Fish and Game Giles W. and El ise G. Mead Foundation We appreciate the generous funding support toward the publication of this book by the United Sta tes Environme ntal Protection Agency, Region IX, San Francisco Contents Pre(acc ix Salmon and Trout, Salmonidae 242 Ackl10 11'ledgl11 el1ts Xlll Silversides, Atherinopsidae 307 COlll'er,<iol1 ['actors xv -

Humboldt Bay Fishes
Humboldt Bay Fishes ><((((º>`·._ .·´¯`·. _ .·´¯`·. ><((((º> ·´¯`·._.·´¯`·.. ><((((º>`·._ .·´¯`·. _ .·´¯`·. ><((((º> Acknowledgements The Humboldt Bay Harbor District would like to offer our sincere thanks and appreciation to the authors and photographers who have allowed us to use their work in this report. Photography and Illustrations We would like to thank the photographers and illustrators who have so graciously donated the use of their images for this publication. Andrey Dolgor Dan Gotshall Polar Research Institute of Marine Sea Challengers, Inc. Fisheries And Oceanography [email protected] [email protected] Michael Lanboeuf Milton Love [email protected] Marine Science Institute [email protected] Stephen Metherell Jacques Moreau [email protected] [email protected] Bernd Ueberschaer Clinton Bauder [email protected] [email protected] Fish descriptions contained in this report are from: Froese, R. and Pauly, D. Editors. 2003 FishBase. Worldwide Web electronic publication. http://www.fishbase.org/ 13 August 2003 Photographer Fish Photographer Bauder, Clinton wolf-eel Gotshall, Daniel W scalyhead sculpin Bauder, Clinton blackeye goby Gotshall, Daniel W speckled sanddab Bauder, Clinton spotted cusk-eel Gotshall, Daniel W. bocaccio Bauder, Clinton tube-snout Gotshall, Daniel W. brown rockfish Gotshall, Daniel W. yellowtail rockfish Flescher, Don american shad Gotshall, Daniel W. dover sole Flescher, Don stripped bass Gotshall, Daniel W. pacific sanddab Gotshall, Daniel W. kelp greenling Garcia-Franco, Mauricio louvar -

Programmatic Essential Fish Habitat (EFH) Assessment for the Long-Term Management Strategy for the Placement of Dredged Material in the San Francisco Bay Region
Programmatic Essential Fish Habitat (EFH) Assessment for the Long-Term Management Strategy for the Placement of Dredged Material in the San Francisco Bay Region July 2009 Executive Summary Programmatic Essential Fish Habitat (EFH) Assessment for the Long-Term Management Strategy for the Placement of Dredged Material in the San Francisco Bay Region Pursuant to section 305(b)(2) of the Magnuson-Stevens Fishery Conservation and Management Act of 1976 (16 U.S.C. §1855(b)), the United States Army Corps of Engineers (USACE) and the United States Environmental Protection Agency (USEPA), as the federal lead and co-lead agencies, respectively, submit this Programmatic Essential Fish Habitat (EFH) Assessment for the Long-Term Management Strategy for the Placement of Dredged Material in the San Francisco Bay Region. This document provides an assessment of the potential effects of the on-going dredging and dredged material placement activities of all federal and non-federal maintenance dredging projects in the action area (see Figure 1.1 located on page 3). The SF Bay LTMS program area spans 11 counties, including: Marin, Sonoma, Napa, Solano, Sacramento, San Joaquin, Contra Costa, Alameda, Santa Clara, San Mateo and San Francisco counties. It does not include the mountainous or inland areas far removed from navigable waters. The geographic scope of potential impacts included in this consultation (action area) comprises the estuarine waters of the San Francisco Bay region, portions of the Sacramento-San Joaquin Delta (Delta) west of Sherman Island and the western portion of the Port of Sacramento and Port of Stockton deep water ship channels. It also includes the wetlands and shallow intertidal areas that form a margin around the Estuary and the tidal portions of its tributaries. -

MARTIN GRIFFIN an Oral History Interview Conducted by Debra Schwartz in 2015
Mill Valley Oral History Program A collaboration between the Mill Valley Historical Society and the Mill Valley Public Library MARTIN GRIFFIN An Oral History Interview Conducted by Debra Schwartz in 2015 © 2015 by the Mill Valley Public Library TITLE: Oral History of Martin Griffin INTERVIEWER: Debra Schwartz DESCRIPTION: Transcript, 37 pages INTERVIEW DATE: October 20th, 2015 In this oral history, physician, naturalist, champion of open spaces and bane of developers Martin Griffin recounts with warmth and humor his long and extraordinarily active life. Born in Ogden, Utah, in 1920 to nature-loving parents, Martin moved with his family to Portland, Oregon, when the Great Depression hit, and then down to Los Angeles and finally up to Oakland, where he attended elementary school through high school. Martin recalls some early experiences that shaped his love for the environment, including his involvement with the Boy Scouts, where he met the graduate student entomologist Brighton C. “Bugs” Cain, who profoundly inspired him. It was also as a boy that Martin came over to Mill Valley for the first time, making his way by ferry and train, to go hiking on Mt. Tamalpais. He conjures the beautiful vision he had from the ridge that day of white birds down on Bolinas Lagoon, a vision which made such a powerful impression on him and would, years later, feed the flames of his conservationist passion. Martin recounts being involved in ROTC while an undergraduate at U.C. Berkeley, later attending medical school at Stanford, where he got married, and moving over to Marin to begin his medical practice. -

Patterns of Evolution in Gobies (Teleostei: Gobiidae): a Multi-Scale Phylogenetic Investigation
PATTERNS OF EVOLUTION IN GOBIES (TELEOSTEI: GOBIIDAE): A MULTI-SCALE PHYLOGENETIC INVESTIGATION A Dissertation by LUKE MICHAEL TORNABENE BS, Hofstra University, 2007 MS, Texas A&M University-Corpus Christi, 2010 Submitted in Partial Fulfillment of the Requirements for the Degree of DOCTOR OF PHILOSOPHY in MARINE BIOLOGY Texas A&M University-Corpus Christi Corpus Christi, Texas December 2014 © Luke Michael Tornabene All Rights Reserved December 2014 PATTERNS OF EVOLUTION IN GOBIES (TELEOSTEI: GOBIIDAE): A MULTI-SCALE PHYLOGENETIC INVESTIGATION A Dissertation by LUKE MICHAEL TORNABENE This dissertation meets the standards for scope and quality of Texas A&M University-Corpus Christi and is hereby approved. Frank L. Pezold, PhD Chris Bird, PhD Chair Committee Member Kevin W. Conway, PhD James D. Hogan, PhD Committee Member Committee Member Lea-Der Chen, PhD Graduate Faculty Representative December 2014 ABSTRACT The family of fishes commonly known as gobies (Teleostei: Gobiidae) is one of the most diverse lineages of vertebrates in the world. With more than 1700 species of gobies spread among more than 200 genera, gobies are the most species-rich family of marine fishes. Gobies can be found in nearly every aquatic habitat on earth, and are often the most diverse and numerically abundant fishes in tropical and subtropical habitats, especially coral reefs. Their remarkable taxonomic, morphological and ecological diversity make them an ideal model group for studying the processes driving taxonomic and phenotypic diversification in aquatic vertebrates. Unfortunately the phylogenetic relationships of many groups of gobies are poorly resolved, obscuring our understanding of the evolution of their ecological diversity. This dissertation is a multi-scale phylogenetic study that aims to clarify phylogenetic relationships across the Gobiidae and demonstrate the utility of this family for studies of macroevolution and speciation at multiple evolutionary timescales. -

DINÂMICA EVOLUTIVA DO Dnar EM CROMOSSOMOS DE PEIXES DA FAMÍLIA ELEOTRIDAE E REVISÃO CITOGENÉTICA DA ORDEM GOBIIFORMES(OSTEICHTHYES, TELEOSTEI)
DINÂMICA EVOLUTIVA DO DNAr EM CROMOSSOMOS DE PEIXES DA FAMÍLIA ELEOTRIDAE E REVISÃO CITOGENÉTICA DA ORDEM GOBIIFORMES(OSTEICHTHYES, TELEOSTEI) SIMIÃO ALEFE SOARES DA SILVA ________________________________________________ Dissertação de Mestrado Natal/RN, Fevereiro de 2019 UNIVERSIDADE FEDERAL DO RIO GRANDE DO NORTE CENTRO DE BIOCIÊNCIAS PROGRAMA DE PÓS-GRADUAÇÃO EM SISTEMÁTICA E EVOLUÇÃO DINÂMICA EVOLUTIVA DO DNAr EM CROMOSSOMOS DE PEIXES DA FAMÍLIA ELEOTRIDAE E REVISÃO CITOGENÉTICA DA ORDEM GOBIIFORMES (OSTEICHTHYES, TELEOSTEI) Simião Alefe Soares da Silva Dissertação apresentada ao Programa de Pós-Graduação em Sistemática e Evolução da Universidade Federal do Rio Grande do Norte, como parte dos requisitos para obtenção do título de Mestre em Sistemática e Evolução. Orientador: Dr. Wagner Franco Molina Co-Orientador: Dr. Paulo Augusto de Lima Filho Natal/RN 2019 Universidade Federal do Rio Grande do Norte - UFRN Sistema de Bibliotecas - SISBI Catalogação de Publicação na Fonte. UFRN - Biblioteca Central Zila Mamede Silva, Simião Alefe Soares da. Dinâmica evolutiva do DNAr em cromossomos de peixes da família Eleotridae e revisão citogenética da ordem gobiiformes (Osteichthyes, Teleostei) / Simião Alefe Soares da Silva. - 2019. 69 f.: il. Dissertação (mestrado) - Universidade Federal do Rio Grande do Norte, Centro de Biociências, Programa de Pós-Graduação em 1. Evolução cromossômica - Dissertação. 2. Diversificação cariotípica - Dissertação. 3. Rearranjos cromossômicos - Dissertação. 4. DNAr - Dissertação. 5. Microssatélites - Dissertação. I. Lima Filho, Paulo Augusto de. II. Molina, Wagner Franco. III. Título. RN/UF/BCZM CDU 575:597.2/.5 SIMIÃO ALEFE SOARES DA SILVA DINÂMICA EVOLUTIVA DO DNAr EM CROMOSSOMOS DE PEIXES DA FAMÍLIA ELEOTRIDAE E REVISÃO CITOGENÉTICA DA ORDEM GOBIIFORMES (OSTEICHTHYES, TELEOSTEI) Dissertação apresentada ao Programa de Pós-Graduação em Sistemática e Evolução da Universidade Federal do Rio Grande do Norte, como requisitos para obtenção do título de Mestre em Sistemática e Evolução com ênfase em Padrões e Processos Evolutivos.